1. Background
Metabolic syndrome is a group of metabolic characteristics that increase the risk of developing cardiovascular diseases and type 2 diabetes. Obesity plays an extremely important role among all the metabolic features of metabolic syndrome. As a result, the prevalence of metabolic syndrome has steadily increased with the growth of the obesity epidemic (1, 2). Cardiovascular diseases are one of the main causes of death worldwide, and in recent years, they have been spreading especially in developing countries. These diseases have different types, with coronary artery disease being the most important, as it is one of the most common causes of heart attacks (3). Metabolic syndrome increases the risk of type 2 diabetes fivefold, cardiovascular disorders twofold, and heart attacks three to four times (4). Increasing age, inactivity, family history, gestational diabetes, race, non-alcoholic fatty liver disease, polycystic ovary syndrome, and genetic mutations are the risk factors of metabolic syndrome (4, 5). Among the genes involved in metabolic syndrome disorder, APOA5, APOC3, CETP, TCF7L2, FTO, GNB3, CRP, CEBP, PPARγ, ADIPOQ, and DYRK1B can be mentioned (6).
The DYRK1B gene encodes a member of the nuclear protein kinase family located on the long arm of chromosome 19 at position 13.2 (19q13.2), which is responsible for many autosomal dominant heart diseases. Another function of the protein kinase DYRK1B is to stimulate the glucose-6-phosphatase enzyme, where increased activity of this enzyme is associated with elevated fasting glucose levels in diabetic patients. The DYRK1B gene is an enzyme cipher that naturally regulates muscle-to-fat balance and controls stable glucose levels through signaling pathways. Mutations in this gene block the stable glucose-maintaining pathways and cause hyperactivity of fat production in the body. It has been observed that intracellular lipid storage in DYRK1B R102C-expressing cells is significantly higher than in non-mutated DYRK1B-expressing cells (7). Recently, two mutations in the DYRK1B gene have been identified, including H90P and R102C, which are associated with the rare autosomal dominant form of abdominal obesity metabolic syndrome. Individuals with this syndrome are at risk of early onset of cardiovascular disease, hypertension, and diabetes. Several studies have shown that the stimulation of this enzyme by protein kinase DYRK1B is dependent on the amount of this protein kinase (7, 8).
A single nucleotide polymorphism (SNP), also called a SNP, is a difference in a DNA sequence in the genome between individuals of the same biological species or between a pair of chromosomes in the same individual. To identify each SNP, it is given a SNP ID or rsID. SNP is the most common type of genetic variation among humans. In a population, a minimal allele frequency can be assigned to SNPs. Most alleles are found among all communities, but some are specific to one community or only part of the ancestral lineage of that community. Therefore, a SNP allele that is common in a geographic region or racial group may be less common among other species. The DYRK1B gene has a SNP rs450819 variant at position 102, which may have a significant relationship between its mutation and the simultaneous occurrence of risk factors in patients with metabolic syndrome. This polymorphism, which is a type of SNV variant in terms of shape, belongs to the intelligent human species, a type of intron variant in the SHKBP1 gene on chromosome number 19 at position 40580108 and in the vicinity of the DYRK1B gene (8).
2. Objectives
The aim of this study was to investigate the effect of the rs450819 polymorphism on metabolic syndrome in people with the DYRK1B gene mutation.
3. Methods
This case-control study was conducted on 121 people from Kherameh in Fars province, which had previously been the focus of researchers due to the prevalence of cardiovascular diseases, to reveal the relationship between this rs450819 polymorphism and metabolic syndrome and its risk factors. For this purpose, blood samples, questionnaire information, and the results of biochemical tests of these people were used. Among the 121 samples, 72 cases were heterozygous for the R102C mutation of the DYRK1B gene, and 49 cases were normal homozygous (as the control group). It is worth mentioning that the participants in this study expressed their satisfaction with the use of the samples prepared for the research.
To determine the genotype related to the rs450819 SNP, DNA extraction was performed from all samples using a unique DNA extraction kit according to the manufacturer's protocol. To amplify the 287 bp fragment of the rs450819 polymorphism (preparation of multiple regional versions of DNA containing the rs450819 SNP based on the sequence and characteristics of the desired region), appropriate primers were designed using Allele ID 7.5 software, and a PCR reaction was performed on the samples. After ensuring the correctness of the PCR, the PCR product of all the samples was sent to Pishgam Company to determine the rs450819 polymorphism for sequencing. The REVERSE primer was used to read the PCR products. Finally, the sequence results of the samples were analyzed. Statistical analysis was done with the help of the chi-square test and SPSS 25 and Prism 6 software, and a significance level of less than 0.05 was considered.
4. Results
In the allelic analysis, the status of allele C was investigated in homozygous and heterozygous individuals in the DYRK1B gene, as well as in individuals with and without metabolic syndrome. Analysis of the results showed no significant relationship between the C allele and metabolic syndrome. Out of the 121 people studied, 49 were homozygous (AA) and 72 were heterozygous (AG) for the DYRK1B gene (Table 1). Using the two definition criteria of ATP3 and IDF, it was determined how many of these genotypes were affected by metabolic syndrome and how many were not affected.
| DYRK1B and rs450819 | Female | Male |
|---|---|---|
| Homozygote (AA); 49 (40); normal = 27; patient = 22 | ||
| TT, 36 (74) | 27 | 9 |
| TC, 9 (18) | 5 | 4 |
| CC, 4 (8) | 3 | 1 |
| Heterozygote (AG); 72 (60); normal = 43; patient = 29 | ||
| TT, 51 (71) | 38 | 13 |
| TC, 19 (26) | 1 | 1 |
| CC, 2 (3) | 11 | 8 |
| Total; n = 121 (100); normal = 70; patient = 51 | ||
| 121 (100) | 85 | 36 |
a Values are expressed as No. (%).
According to the available findings, the likelihood of observing the C allele as a mutant allele compared to the T allele in the patient group is almost equal to the likelihood of this allele being present in the group of healthy individuals (Table 2).
| Alleles and Genotypes | Normal (N = 70) | Patient (N = 51) |
|---|---|---|
| rs450819 (alleles) | ||
| T, 202 (83.5) | 118 (84.3) | 84 (82.4) |
| C, 40 (16.5) | 22 (15.7) | 18 (17.6) |
| rs450819 (genotypes) | ||
| TT, 87 (72) | 51 (59) | 36 (41) |
| TC, 28 (23) | 16 (57) | 12 (43) |
| CC, 6 (5) | 3 (50) | 3 (50) |
a Values are expressed as No. (%).
Allelic frequency distribution in both the case (heterozygous) and control (homozygous) groups showed that the T allele has the highest frequency in both heterozygous (84%) and homozygous (83%) groups, while the C allele has the lowest frequency in both heterozygous (17%) and homozygous (23%) groups, indicating that the C allele is mutant. In other words, there is no significant difference in allele frequency between the two case-control groups (Tables 3 and 4).
| Variables | Male | Female | ||
|---|---|---|---|---|
| Patient (N = 12) | Normal (N = 24) | Patient (N = 39) | Normal (N = 46) | |
| Allele C | 5 (21) | 11 (23) | 13 (17) | 11 (12) |
| Allele T | 19 (79) | 37 (77) | 65 (83) | 81 (88) |
a Values are expressed as No. (%).
| Variable | T | C |
|---|---|---|
| DYRK1B | ||
| Homozygote | 81 (83) | 17 (17) |
| Heterozygote | 121 (84) | 23 (16) |
a Values are expressed as No. (%).
In the allelic analysis, the status of the C allele in affected and non-affected individuals indicates the absence of a significant relationship between the C allele and metabolic syndrome (Table 5).
| Variable | Patient | Normal |
|---|---|---|
| Allele C | ||
| Homozygote (N = 17) | 10 (58.8) | 7 (41.2) |
| Heterozygote (N = 23) | 8 (34.8) | 15 (65.2) |
a Values are expressed as No. (%).
Subsequently, the genotypic examination (examining the relationship of genotypes with each other and with metabolic syndrome) was conducted using the chi-square test and Prism 6 software, with a significance level of less than 5% considered. The result, similar to the allelic examination, indicates the absence of a relationship between the CC and CT genotypes with other genotypes and with metabolic syndrome. Therefore, the presence of the C allele cannot be considered a factor in increasing the risk of metabolic syndrome, nor can the C allele be defined as a protective allele (Tables 6 and 7).
| Variables | Normal | Patient | Adjusted Odds Ratio (95% CI) | Adjusted P-Value |
|---|---|---|---|---|
| Alleles | 0.8701 (0.4394 - 1.723) | 0.6894 | ||
| C | 22 | 18 | ||
| T | 118 | 84 |
| Variables | Normal | Patient | Adjusted Odds Ratio (95% CI) | Adjusted P-Value |
|---|---|---|---|---|
| Allele | 1.069 (0.3882 - 2.941) | 0.8978 | ||
| C | 15 | 8 | ||
| T | 71 | 50 |
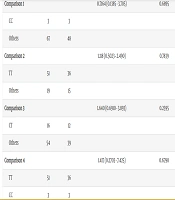
Analysis of data regarding this polymorphism, as well as genotypic and allelic data, showed that there is no significant relationship between the presence of the C allele and the risk of metabolic syndrome (Table 8).
| Genotypes | Normal | Patient | Adjusted Odds Ratio (95% CI) | Adjusted P-Value |
|---|---|---|---|---|
| Comparison 1 | 0.7164 (0.1385 - 3.705) | 0.6895 | ||
| CC | 3 | 3 | ||
| Others | 67 | 48 | ||
| Comparison 2 | 1.118 (0.5023 - 2.490) | 0.7839 | ||
| TT | 51 | 36 | ||
| Others | 19 | 15 | ||
| Comparison 3 | 1.640 (0.6910 - 3.893) | 0.2595 | ||
| CT | 16 | 12 | ||
| Others | 54 | 39 | ||
| Comparison 4 | 1.417 (0.2703 - 7.425) | 0.6790 | ||
| TT | 51 | 36 | ||
| CC | 3 | 3 |
5. Discussion
Today, metabolic syndrome is an important clinical problem worldwide. Despite progress in this field, prevention remains the best way to address this disorder, making the identification of its risk factors very important. Additionally, due to the association of this syndrome with the occurrence of cardiovascular diseases, patients with this syndrome are prioritized for interventional measures to prevent coronary artery diseases (2, 9). Genetic factors, such as the R102C mutation in the DYRK1B gene, as well as several polymorphisms, have been identified as increasing the risk of this disease. Identification of genetic polymorphisms, including SNPs, is very helpful for diseases, especially those resulting from the action of multiple genes. Disease-related polymorphisms are usually identified through case-control studies and repeated studies in different populations, and they are confirmed as indicators for predicting or identifying the disease (10).
With the progress of modern molecular genetics, various research has been conducted to determine the genetic factors of metabolic syndrome. The most progress has been made in identifying common variants that increase the risk of one or two metabolic risk factors (11). In the history of similar studies, many have proven the effect of different variants on the incidence of metabolic syndrome. Numerous studies have been conducted on genes and genetic factors affecting metabolic syndrome. In a systematic comprehensive study on genes that had at least one associated SNP, it was observed that 8 SNPs were most associated with metabolic syndrome, most of which were related to genes associated with fat metabolism. Following these studies, many investigations have been conducted on SNPs related to metabolic syndrome or one or more of its risk factors in different communities (12).
In similar studies on the DYRK1B gene variant, the L28P variant was found to have a protective effect against metabolic syndrome (13). The investigation of the association of rs10499859 A > G and rs12346514 C > T polymorphisms of the CD36 gene on chromosome 7 with metabolic syndrome showed that these SNPs were associated with metabolic syndrome indicators such as HDL cholesterol and Body Mass Index. They are related, although considering the participation of many environmental and genetic factors in the occurrence of metabolic syndrome, the role of each of these polymorphisms alone is small (14).
In another study, the relationship between the rs2236242 polymorphism of the VASPIN gene (TT-TA) and the duration of walking and metabolic syndrome and coronary artery disease was investigated. It was found that walking longer than 60 minutes per day significantly reduces the metabolic risk of the syndrome and its components, such as blood pressure and triglyceride concentration, in TA carriers (15).
The findings of recent research on the relationship between the rs450819 polymorphism related to the DYRK1B gene and the occurrence of metabolic syndrome did not show a relationship between this polymorphism and metabolic syndrome. The statistical analysis of the results showed that the mutation of this SNP has no effect on the exacerbation of metabolic syndrome, and its activity was not significantly different between the patient and control groups. In other words, no significant difference was observed in the allelic distribution of this polymorphism between the two patient and control groups. The analysis of the data on the rs450819 polymorphism showed that there is no significant relationship between the CC genotype and the risk of metabolic syndrome. Also, the allelic analysis showed that there is no significant relationship between the C allele and the risk of metabolic syndrome.
However, in similar investigations for the rs13246513 polymorphism of the CD36 gene, it was observed that this SNP increases the risk of metabolic syndrome by 32% (16). The difference in the findings could be related to the different genetic backgrounds or the number of samples in the study population.