Abstract
Context:
Medical imaging technologies are an indispensable tool in medicine today developed to satisfy the significant demand for information on medical imaging by visualizing internal organs for clinical analysis. This enables the radiologists and clinicians to accurately understand the patient’s condition and makes medical practices easier, more effective for patients, and cheaper for the healthcare system.Objective:
The current study aimed at presenting a comprehensive review on the recent classification and segmentation techniques of brain tumors in magnetic resonance image (MRI).Data Source:
Google Scholar, ScienceDirect, Web of Knowledge, Springer, and manual search of reference lists from 1990 to 2018.Inclusion Criteria:
The current study considered brain tumors since they are relatively less common and more important compared with other tumors due to their high morbidity rate.Results:
Many automated brain tumors segmentation algorithms of magnetic resonance imaging (MRI) were reviewed and discussed including their advantages and limitations to provide a clear insight into these algorithms. The review concentratedon the state-of-art methods of segmentation of MRI brain tumors since they attracted a significant attention in the recent two decades resulting in many algorithms being developed for automated, semi-automated, and interactive segmentation of brain tumors. While there is a significant development of segmentation algorithms, they are rarely used clinically due to lack of interaction between developers and clinicians.Conclusions:
Most studies did not consider grading of brain tumors and did not distinguish to which grade the brain tumor belonged. This enables the developers to understand how the margins of brain tumors appear in medical images.Limitations:
The most important limitations that make brain tumors segmentation remaina challenging task are the variety of the shape and intensity of tumors in addition to the probability of inhomogeneity of tumorous tissue.Keywords
Classification Segmentation Magnetic Resonance Imaging Brain Tumors
1. Context
The medical imaging technologies revolutionized medical diagnosis over the last 40 years allowing doctors to detect tumors earlier and improve the prognosis. Moreover, they give physicians the ability to investigate the internal structures and functions of the human body with a range of imaging systems and use them to plan treatments and surgeries (1, 2). Typical medical imaging technologies include ultrasonography (US), computed tomography (CT), and magnetic resonance imaging (MRI). MRI is the most commonly used system to diagnose brain tumors since it presents accurate details such as the type, position, and size of the investigated tumor. Additionally, it is capable of differentiating soft tissue with high resolution and is more sensitive detecting and visualizing subtle changes in tissue density and the physiological alternations associated with the tumor (3-7). MRI has further role to help the clinicians move to precise lesion diagnosis instead of the indirect diagnosis using cerebral angiography (8). Furthermore, MRI is different from the other medical technologies due to its capability to use different image acquisition protocols to produce multiple images with different contrast visualization of the same tissue. These protocols may slightly vary and provide more valuable anatomical information to help the clinicians study the diseased brains precisely (3, 5, 8, 9).
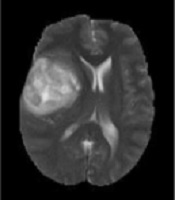
Generally, based on the advantages of the MRI, modalities can be categorized into T1-w images, which are anatomical images and beneficial for black hole detection, which looks as hypo-intense or dark area relative to the white matter (WM) intensities. On the other hand, T2-w images are suitable for tissue pathology and show well-defined tumor delineation. Moreover, the WM lesions are shown as hyper-intense or bright areas relative to the WM intensities. Therefore, T2-w images are particularly useful for pathological detection (10). The main drawback of this modality is that the cerebrospinal fluid (CSF), grey matter (GM), and tumors have close intensities (9). Clinically, T2-w and T1c-w are the first choice of brain tumor diagnosis methods, but using these two MRI modalities can produce difficulties to differentiate between the new and old tumors or tumors from non-tumoral lesions in addition to grading (6). The analysis of such types of MR images requires advanced computerized tools as well as digital image processing technology. Sometimes, using a contrast enhancement material is essential to clearly highlight the edges of a brain tumor in T1-w images. This is important to distinguish and recognizesome types of brain tumors in T2-w and T1-w images (5, 11, 12). Figure 1 shows samples of T2-w, T1-w, Fluid-attenuated inversion recovery (FLAIR), and T1c-w pathological slices.
One of the special challenges in MR images is the brain tumors identification since the existence of a brain tumor in MR images can be linked to a highly inhomogeneous signal that can be related to the signal strength of the normal tissue (6, 7, 13, 14). Ambiguity in classification of pixels within the tumor region can lead to inaccurate segmentation and it occurs when some parts of the tumor cannot be distinguished from WM/GM due to the limited intensity resolution of the MR image and the complexity of the human brain anatomy. This happens at the boundary of brain tumor and surrounding normal tissue as a result of the influence of partial volumes (PV) (15). Therefore, the PV blurs the MR images so much and leads to mixing in the intensity value of each voxel with its neighbors (3, 12, 16).
The current study aimed at providing a review of the automated brain tumors segmentation algorithms and presenting a thorough analysis of these algorithms. The rest of the study was organized as follows: In sections 2 and 3, a summery about brain tumors segmentation and conclusion are demonstrated, respectively.
Samples of four pathological MRI slices (T2-w, T1-w, FLAIR, and T1c-w) from left to right

2. Brain Tumors Segmentation
There is a variety of medical imaging technologies employed to help the clinicians identify pathological conditions inside the body, congenital defects, functionality of the organs and vessels, broken bones, and tumors (17). The increasing number of medical imaging technologies and massiveness of clinical data generation made it impossible to manually classify and segment the data in short time (17). Therefore, computer algorithms are employed to help in specific tasks such as the detection and classification of tumors (18). The computer applications that support medical imaging techniques use image processing algorithms for quantitative analysis to help clinicians who are currently assessing and diagnosing medical images visually. However, these applications have some limitations in terms of time and accuracy. The reasons behind these limitations are inter-observer variations and error due to stress, oversight, and limited experience. Hence, computer analysis provides great supports that can help for the subjective diagnosis, and thus, it is essential to improve diagnostic accuracy and confidence even for experts with high experience.
Image segmentation is image processing of partitioning the input image into separate areas containing similar pixels attributes. Extensive different brain tumor segmentation techniques are recently proposed due to quick progress in the medical imaging technology (19, 20). In general, segmentation techniques are classified based on the image information employed to implement the segmentation.
2.1. Pixel Based Segmentation
This type of segmentation is also known as threshold-based methods. They are conceptually the simplest segmentation approach and commonly used in two-dimensional images. They only consider the intensity value of the current pixel and discard its neighboring pixels. Most of pixel based techniques essentially depend on measuring thresholds from the histogram of an image (21-24). If the object can be segmented by a single threshold, it is noted as global thresholding. However, if there are more than two objects, then the segmentation should be implemented using local thresholding (20, 23, 25, 26). The main problem of this type of segmentation is that only the intensity information is considered and the relationships between the pixels are neglected; therefore, some pixels do not attend the desired or the background regions. In general, the threshold-based segmentation methods failed to exploit the provided information by MRI slice and in most cases are usually used to separate and eliminate the background of MRI slice (20, 26, 27).
2.2. Region Based Segmentation
This approach is based on dividing the image into regions according to predefined similarity criteria. It is also called region merging and starts with a single pixel or a group of pixels called seeds. Neighbors of the seeds are checked and only the pixels that satisfy the similarity criteria to the same structure of interest are added (28). The similarity between pixels can be based on intensity information and/or edges in the image (18). The procedure is repeated until no more pixels are added to the structure of interest. The main characteristic of region growing method is the capability to segment similar regions and generate related regions (29). The main disadvantage of region growing methods is the PV effect, which limits the accuracy of MR brain image segmentation. Therefore, PV blurs the borders between different tissue, since the voxel may contain more than one kind of tissue types (20, 25, 30). Region growing methods are more sensitive to noise, thus producing holes in the extracted regions (18). Additionally, if the seed point is not properly chosen, the region grows outside the object of interest or merges with another region that does not belong to the desired object (22).
2.3. Edge-Based Segmentation
Edge-based segmentation is based on finding the differences (instead of similarities) between pixels to determine the close boundaries corresponding to the objects of an image (22). Edge-based segmentation is computationally fast and does not need any prior information about image content (29). It is developed to be strongly sensitive to the significant variations in grey level values, and determines if a pixel lies on an edge independently (2). This approach can be used to overcome the effect of changing the size of the segmented object due to the unsuitable thresholding scheme used for segmentation (31). The main limitation of edge-based segmentation is that the resulting edges do not enclose the object completely. To solve this problem, extra post-processing steps should be taken to link edges that correspond to a single boundary in order to combine these edges into chains to improve the representing edges in the image. They are more sensitive to image noise, and if the images of the region features differ by only a small quantity between regions, detected edges may be broken (29, 32, 33).
2.4. Deformable Model
The contour or snake models are types of the parametric deformable models, which are suitable to segment, match, and track the pathological structures in MR images by exploiting the derived constraints from MR images, and prior knowledge about the tumor location, size, and shape of these pathological areas (20, 22, 31). These deformable models are defined as a set of curves directed by the impact of internal and external forces. The effect of internal forces smooths the curves, while external forces are responsible to change the direction of the curves toward the edges of anatomical area. Among all segmentation techniques, the deformable model was a successful and efficient technique. The deformable model is used for a wide range of applications, especially in medical fields, due to its capability to accommodate the variability of biological structures of different patients (10, 34). Jin et al. (25), and Gordillo et al. (20), concluded that the good results of brain tumor segmentation using conventional methods (e.g., region based method, pixel based method, and edge based method) are hard to achieve. Additionally, due to the emersion of volumetric three-dimensional medical imaging data, the segmentation of this data is a challenging problem to extract the boundary features that belong to the same structure.
Previous studies commonly focused on segmenting each slice individually (slice-by-slice), then merging them to obtain a three-dimensional volume or a continuous surface. However, the resulting segmentation leads to a non-continuous surface since some important anatomical information of the full MRI slices is missing (35, 36). A three-dimensional deformable model approach was used as the best segmentation method that does not need training data. It also requires initializing the contour that is close to the object of interest. The approach also employs user guidance to place landmarks in the image to steer the segmentation (37). The main advantage of the deformable model approaches is the ability to extract boundary featuresfor the same regions (25, 34).
2.5. Machine-Learning-Based Segmentation
Machine learning is a one of the most effective methods to automate the analysis and segmentation of medical images. It can learn the available complex relationships from the empirical data to make accurate decisions (25). Machine-learning-based approaches for image segmentation are classified into three categories: Supervised, semi-supervised, and unsupervised segmentation methods. Once the training data is manually labelled, the segmentation method is said to be supervised. The main advantage of supervised segmentation method is that it is possible to use them to perform different tasks by only changing the training set (20). If the training data are automatically labelled by numerically grouping similar pixels, the segmentation is said to be unsupervised (20, 28, 38, 39). This type of segmentation uses intensities and/or texture features to segment MRI scan into remarkable regions. The presence of more than 120 brain tumors with different shapes and intensities makes achieving accurate segmentation of brain tumors more complicated and challenging, especially when the intensities of the tumors are heterogeneous and they have unclear boundaries (20, 40). The majority of brain tumor segmentation algorithms are based on artificial neural networks (ANN), support vector machine (SVM), and fuzzy C-means clustering (FCM).
ANN technique is a one of the most popular segmentation methods because of its ability to learn from historical cases and automatically generate new rules (41). ANN uses intensities and texture features to segment the MRI brain scan. The extracted features are fed through a series of nodes named input layers and then a set of mathematical operations are applied to these features within a hidden layer. The final decision is made at the output layer (20). No standard approach is used to identify the network structure that represents the best for brain tumor segmentation. Hence, tedious experiments and trial and error are often used (42). These limitations are overcome using convolutional neural network (CNN) (41, 43).
The support vector machine (SVM) was developed in 1992 by Vapnik et al. (44). The SVM is considered as one of the supervised learning models used in various applications such as segmentation, object recognition, speaker identification, and medical diagnosis (10, 38). It was used to classify voxels into normal and pathological tissue (45-47).
FCM is an unsupervised technique of segmenting and grouping pixels into two or more clusters by generating a set of memberships where each one covers a group of pixels corresponding to the nearest cluster center (25). Therefore, the assigned membership of a specific class is determined according to its characteristics such as intensity values and texture features, and it can be accurately determined through the estimation of cluster centers (20). The main disadvantages of the FCM are that it is very time consuming, and highly sensitive to the noise and inhomogeneity, which leads to erroneous segmentation results (48).
2.6. Atlas-Based Segmentation
Atlas-based approaches are used significantly in medical image segmentation and are widely employed in computer aided diagnosis to determine the object shapes or detect the morphological differences between patient groups (49). The atlas brings useful prior knowledge about the brain anatomy used as a reference to segment new MR images. This enables the segmentation of any brain tumors available in the atlas without any additional cost (36). The atlas-based segmentation technique remains challenging and is yet to be used in general applications since it is based on training sets (45, 50). Furthermore, the atlas registration introduces a bias in the segmentation since the algorithm searches for shape similarity to one of the atlases (34). In addition, atlas requires more time to construct, which in turn represents the main drawback of this approach (40). Details of the current review study such as advantages and disadvantages are formulated in Table 1.
The Advantages and Disadvantages of Brain Tumor Segmentation Methods
| Reference | Method | Advantages | Disadvantages |
|---|---|---|---|
| Nabizadeh (50) | Segmentation-based texture features (first-order statistical, GLCM (4 orientations and 2 distances), GLRLM (4 orientations), histogram of gradient (HOG), LBP, anisotropic Complex Morlet Wavelet transform) and SVM classifier | (1) Independent of atlas registration, (2) Independent of prior anatomical knowledge, (3) Independent of bias correlation, (4) Using single-spectral MRI | High computational complexity |
| Al-Waeli (3) | Three-dimensional active contour without edge (3DACWE) | (1) This approach does not consider the local tumor properties (gradients), global properties (intensity), contour length, and region length, (2) This approach does not rely on atlas registration and the prior anatomical knowledge, (3) It does not need to initialize assumptions about the number of classes in MRI scan. | High computational complexity |
| Ibrahim et al. (51) | Deformable model based on fractional Wright energy function (FWF) | (1) The proposed FWF method minimized the energy function more than the gradient-decent method that was used in the original three-dimensional active contour without edge (3DACWE) method, (2) The proposed 3DACWE with FWF method offers a high accuracy compared with that of the original 3DACWE method, | High computational complexity |
| Sachdeva et al. (52) | Content-based active contour (CBAC) | (1) It is a semi-automatic segmentation method, where the initial seed point is chosen by the radiologist, (2) It considers both intensity and texture inside the pathological area, (3) This approach segments the tumor boundaries regardless of the heterogeneity, and weak and false edges. | - |
| Guo et al. (47) | The proposed system used an unsupervised and a supervised component. In the unsupervised component, the pathological hemisphere was identified and in supervised component the segmentation-based texture features (zero-order statistical, first-order statistical, second order statistical) and SVM classifier were implemented. | Fully-automated system | (1) The partial volume, which affects the performance of the method, (2) The similarity in intensities between the lesion and CSF of brain, (3) Bias field inhomogeneity, which affects the identification of the lesioned hemisphere, (4) When both hemispheres contain lesions |
| Sanjuan et al. (13) | (1) A modified implementation of the unified segmentation-normalization procedure of SPM, (2) Fuzzy-logic clustering method | (1) It is able to recognize brain tumors at the correct location in all pathological patients irrespective of the type, size, and location, (2) It does not care that the tumor appears brighter or darker in MRI images, (3) Fully automated system. | (1) It failed to recognize high grade brain tumors, (2) It failed to identify tiny brain tumors (few millimeters). |
| Soltaninejad et al. (53) | It is based on super-pixel technique and classification of each super- pixel. The considered feature techniques were intensity-based, Gabor textons, fractal analysis and curvatures, and extremely randomized trees (ERT) classifier. | Fully automated system | (1) It is not suitable for small-sized lesions, (2) The computation time to generate the small size partitions of super-pixel is very high, (3) Time consuming |
| Havaei et al. (43) | It is based on deep neural networks (DNNs). | (1) It does not need to implement pre-processing algorithms, (2) This approach has efficiently extracted the complex features, (3) It has less outlier than other proposed approaches. | It requires implementation of post-processing algorithm to remove flat holes that might appear in the segmented image. |
| Zhang et al. (54) | It is based on using fully convolutional neural network (FCNN), bootstrapping loss, dice loss and sensitivity-specificity loss. | It shows powerful and efficient distinguishing ability compared with the original design of CNN. | (1) It gives more false positive predictions than expected while classifying enhancing tumor, (2) The model also fails to give boundaries between classes as fine as the ground truth images, (3) Huge memory demand makes a trade-off between accuracy and resource consumption. |
| Kahali et al. (48) | It is based on using modified fuzzy c-means algorithm (MoFCM) and followed by modified spatial fuzzy c-means (MSFCM). | It is less sensitive to the generated noise and the intensity of inhomogeneity. | - |
| De et al. (55) | It is based on using fuzzy inter-cluster hostility index-based GA method | (1) It does need any prior information, (2) It is an unsupervised method | - |
| Lee et al. (56) | It is based on using the surface evolution principle based on the geometric deformable model and the level set theory | (1) It converged faster to steady-state with minimum number of iterations, (2) More accurate in segmenting brain tumors, (3) It is less sensitive to the noise in MR images. | - |
3. Conclusions
Accurate segmentation of brain tumors is important for clinical diagnosis, predicting the prognosis, and treatment. It is beneficial for the general modeling of pathological brain topology and the exploration of the anatomical construction of the brain and any tumors it may contain. In the current study, many automated brain tumors segmentation algorithms of MRI were reviewed and discussed including their advantages, and limitations to provide a clear insight into such algorithms. The review concentrated on the state-of-art methods of segmentation of MRI brain tumors since they attracted a significant attention in recent two decades, resulting in many algorithms being developed for automated, semi-automated, and interactive segmentation of brain tumors. While there was a significant development of segmentation algorithms, they were rarely used clinically due to lack of interaction between developers and clinicians. Although there are many existing brain tumor segmentation algorithms, manual segmentation is preferred clinically due to the lack of interpretability and easy handling of the automatic segmentation tools. Many factors should be considered to improve the confidence of automatic segmentation tools such as being more user-friendly, robust, and accurate.
Acknowledgements
References
-
1.
Bankman IN. Handbook of medical image processing and analysis. 2th ed. Burlington: Elsevier/Academic Press; 2009.
-
2.
Birry RAK. Automated classification in digital images of osteogenic differentiated stem cells [dissertation]. University of Salford; 2013.
-
3.
Al-Waeli AMH. An automated system for the classification and segmentation of brain tumours in MRI images based on the modified grey level co-occurrence matrix [dissertation]. Salford, UK: University of Salford; 2017.
-
4.
Loizou CP. Ultrasound image analysis of the carotid artery [dissertation]. London, UK: Kingston University; 2005.
-
5.
Drevelegas A, Papanikolaou N. Imaging modalities in brain tumors. In: Antonios D, editor. Imaging of brain tumors with histological correlations. Springer Berlin Heidelberg; 2011. p. 13-33.
-
6.
Tonarelli L. Magnetic resonance imaging of brain tumor. 300. Enterprises for Continuing Education, Inc; 2013. p. 48116-300.
-
7.
Mechtler L. Neuroimaging in neuro-oncology. Neurol Clin. 2009;27(1):171-201. ix. [PubMed ID: 19055979]. https://doi.org/10.1016/j.ncl.2008.09.015.
-
8.
Strong M, Garces J, Vera J, Mathkour M, Emerson N, Ware ML. Brain tumors: Epidemiology and current trends in treatment. J Brain Tumor Neurooncol. 2015;1(1). https://doi.org/10.4172/jbtn.1000102.
-
9.
Mortazavi D, Kouzani AZ, Soltanian-Zadeh H. Segmentation of multiple sclerosis lesions in MR images: A review. Neuroradiology. 2012;54(4):299-320. [PubMed ID: 21584674]. https://doi.org/10.1007/s00234-011-0886-7.
-
10.
Tantisatirapong S. Texture analysis of multimodal magnetic resonance images in support of diagnostic classification of childhood brain tumours [dissertation]. University of Birmingham, UK; 2015.
-
11.
Belkic D, Belkic K. Signal processing in magnetic resonance spectroscopy with biomedical applications. CRC Press; 2010.
-
12.
Hasan A, Meziane F, Aspin R, Jalab H. Segmentation of brain tumors in MRI images using three-dimensional active contour without edge. Symmetry. 2016;8(11):132. https://doi.org/10.3390/sym8110132.
-
13.
Sanjuan A, Price CJ, Mancini L, Josse G, Grogan A, Yamamoto AK, et al. Automated identification of brain tumors from single MR images based on segmentation with refined patient-specific priors. Front Neurosci. 2013;7:241. [PubMed ID: 24381535]. [PubMed Central ID: PMC3865426]. https://doi.org/10.3389/fnins.2013.00241.
-
14.
Pantelis G. Computer assisted diagnosis of brain tumors based on statistical methods and pattern recognition techniques [dissertation]. University of Patras, Greece; 2010.
-
15.
Tohka J. Partial volume effect modeling for segmentation and tissue classification of brain magnetic resonance images: A review. World J Radiol. 2014;6(11):855-64. [PubMed ID: 25431640]. [PubMed Central ID: PMC4241492].
-
16.
Zhang Y, Brady JM, Smith SM. An HMRF-EM algorithm for partial volume segmentation of brain MRI FMRIB technical report TR01YZ1. Department of Engineering Science, Centre for Functional Magnetic Resonance Imaging of the Brain, Oxford University; 2001.
-
17.
Mohan G, Subashini MM. MRI based medical image analysis: Survey on brain tumor grade classification. Biomed Signal Proces. 2018;39:139-61. https://doi.org/10.1016/j.bspc.2017.07.007.
-
18.
Palm DL, Xu C, Prince JL. A survey of current methods in medical image segmentation. Baltimore, USA: Technical report, Johns Hopkins University; 1998.
-
19.
Menze BH, Jakab A, Bauer S, Kalpathy-Cramer J, Farahani K, Kirby J, et al. The multimodal brain tumor image segmentation benchmark (BRATS). IEEE Trans Med Imaging. 2015;34(10):1993-2024. [PubMed ID: 25494501]. [PubMed Central ID: PMC4833122]. https://doi.org/10.1109/TMI.2014.2377694.
-
20.
Gordillo N, Montseny E, Sobrevilla P. State of the art survey on MRI brain tumor segmentation. Magn Reson Imaging. 2013;31(8):1426-38. [PubMed ID: 23790354]. https://doi.org/10.1016/j.mri.2013.05.002.
-
21.
Petrou M. Texture in biomedical images. In: Deserno MT, editor. Biomedical image processing. Springer Berlin Heidelberg; 2010. p. 157-76. https://doi.org/10.1007/978-3-642-15816-2_6.
-
22.
Dougherty G. Digital image processing for medical applications. Cambridge University Press; 2009.
-
23.
Naji SA, Zainuddin R, Jalab HA. Skin segmentation based on multi pixel color clustering models. Digit Signal Process. 2012;22(6):933-40. https://doi.org/10.1016/j.dsp.2012.05.004.
-
24.
Alan H. W, Policarpo F. The computer image. the University of Michigan: Addison-Wesley; 1998.
-
25.
Jin L, Min L, Jianxin W, Fangxiang W, Tianming L, Yi P. A survey of MRI-based brain tumor segmentation methods. Tsinghua Sci Technol. 2014;19(6):578-95. https://doi.org/10.1109/tst.2014.6961028.
-
26.
Russ JC. Image processing. Computer-assisted microscopy. Springer; 1990. p. 33-69.
-
27.
Wilson JN, Ritter GX. Handbook of computer vision algorithms in image algebra. CRC Press; 2000.
-
28.
Solomon C, Breckon T. Fundamentals of digital image processing: A practical approach with examples in matlab. John Wiley & Sons; 2011.
-
29.
Rogowska J. Chapter 5- Overview and fundamentals of medical image segmentation. In: Bankman IN, editor. Handbook of medical image processing and analysis. Elsevier/Academic Press; 2009.
-
30.
Mie S, Lakare S, Ming W, Kaufman A, Nakajima M. A gradient magnitude based region growing algorithm for accurate segmentation. Proceedings 2000 International Conference on Image Processing (Cat. No.00CH37101). Vancouver, BC, Canada, Canada. IEEE; 2000. p. 448-51.
-
31.
Jähne B. Digital image processing. 60th ed. Berlin: Springer; 2005.
-
32.
Sonka M, Hlavac V, Boyle R. Image processing, analysis, and machine vision. Cengage Learning; 2014.
-
33.
William K. Digital image processing. 3th ed. Canada: Wiley-Interscience; 2001. 656 p.
-
34.
Rousseau O. Geometrical modeling of the heart [dissertation]. Canada: University of Ottawa; 2009.
-
35.
Mikulka J, Gescheidtova E. An improved segmentation of brain tumor, edema and necrosis. Proceedings of Electromagnetics Research Symposium Proceedings,Taipei. 2013. p. 25-8.
-
36.
Despotovic I, Goossens B, Philips W. MRI segmentation of the human brain: Challenges, methods, and applications. Comput Math Methods Med. 2015;2015:450341. [PubMed ID: 25945121]. [PubMed Central ID: PMC4402572]. https://doi.org/10.1155/2015/450341.
-
37.
Loizou CP, Pantziaris M, Seimenis I, Pattichis CS. Brain MR image normalization in texture analysis of multiple sclerosis. 9th International Conference on Information Technology and Applications in Biomedicine, Larnaca. 2009. p. 1-5.
-
38.
Han J, Pei J, Kamber M. Data mining: Concepts and techniques. 3th ed. Elsevier Science; 2011.
-
39.
Larose DT. Discovering knowledge in data: An introduction to data mining. USA: John Wiley & Sons; 2005.
-
40.
Angulakshmi M, Lakshmi Priya GG. Automated brain tumour segmentation techniques: A review. Int J Imag Syst Tech. 2017;27(1):66-77. https://doi.org/10.1002/ima.22211.
-
41.
Negnevitsky M. Artificial intelligence: A guide to intelligent systems. Addison-Wesley; 2005.
-
42.
Zhang G, Eddy Patuwo B, Y. Hu M. Forecasting with artificial neural networks. Int J Forecasting. 1998;14(1):35-62. https://doi.org/10.1016/s0169-2070(97)00044-7.
-
43.
Havaei M, Davy A, Warde-Farley D, Biard A, Courville A, Bengio Y, et al. Brain tumor segmentation with Deep Neural Networks. Med Image Anal. 2017;35:18-31. [PubMed ID: 27310171]. https://doi.org/10.1016/j.media.2016.05.004.
-
44.
Hasan AM, Meziane F. Automated screening of MRI brain scanning using grey level statistics. Comput Electr Eng. 2016;53:276-91. https://doi.org/10.1016/j.compeleceng.2016.03.008.
-
45.
Nabizadeh N, Kubat M. Brain tumors detection and segmentation in MR images: Gabor wavelet vs. statistical features. Comput Electr Eng. 2015;45:286-301. https://doi.org/10.1016/j.compeleceng.2015.02.007.
-
46.
Bauer S, Nolte LP, Reyes M. Fully automatic segmentation of brain tumor images using support vector machine classification in combination with hierarchical conditional random field regularization. International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer Berlin Heidelberg; 2011. p. 354-61.
-
47.
Guo D, Fridriksson J, Fillmore P, Rorden C, Yu H, Zheng K, et al. Automated lesion detection on MRI scans using combined unsupervised and supervised methods. BMC Med Imaging. 2015;15:50. [PubMed ID: 26518734]. [PubMed Central ID: PMC4628334]. https://doi.org/10.1186/s12880-015-0092-x.
-
48.
Kahali S, Adhikari SK, Sing JK. 3D MRI brain image segmentation: A two-stage framework. International Conference on Computational Intelligence, Communications, and Business Analytics. Springer; 2017. p. 323-35.
-
49.
Kalinić H. Atlas-based image segmentation: A survey. Universiy Zagreb. 2009.
-
50.
Nabizadeh N. Automated brain lesion detection and segmentation using magnetic resonance images. University of Miami, USA; 2015.
-
51.
Ibrahim RW, Hasan AM, Jalab HA. A new deformable model based on fractional Wright energy function for tumor segmentation of volumetric brain MRI scans. Comput Methods Programs Biomed. 2018;163:21-8. [PubMed ID: 30119853]. https://doi.org/10.1016/j.cmpb.2018.05.031.
-
52.
Sachdeva J, Kumar V, Gupta I, Khandelwal N, Ahuja CK. A package-SFERCB-“Segmentation, feature extraction, reduction and classification analysis by both SVM and ANN for brain tumors”. Appl Soft Comput. 2016;47:151-67. https://doi.org/10.1016/j.asoc.2016.05.020.
-
53.
Soltaninejad M, Yang G, Lambrou T, Allinson N, Jones TL, Barrick TR, et al. Automated brain tumour detection and segmentation using superpixel-based extremely randomized trees in FLAIR MRI. Int J Comput Assist Radiol Surg. 2017;12(2):183-203. [PubMed ID: 27651330]. [PubMed Central ID: PMC5263212]. https://doi.org/10.1007/s11548-016-1483-3.
-
54.
Zhang J, Shen X, Zhuo T, Zhou H. Brain tumor segmentation based on refined fully convolutional neural networks with a hierarchical dice loss. Comput Vision Pattern Recog. 2017.
-
55.
De S, Bhattacharyya S, Dutta P. Automatic magnetic resonance image segmentation by fuzzy intercluster hostility index based genetic algorithm: An application. Appl Soft Comput. 2016;47:669-83. https://doi.org/10.1016/j.asoc.2016.05.042.
-
56.
Lee M, Cho W, Kim S, Park S, Kim JH. Segmentation of interest region in medical volume images using geometric deformable model. Comput Biol Med. 2012;42(5):523-37. [PubMed ID: 22402196]. https://doi.org/10.1016/j.compbiomed.2012.01.005.