Abstract
Background:
Acute respiratory infection plays a major role in hospitalization of children worldwide and viruses are one of the most important causative pathogens of acute respiratory tract infections (ARTIs). Human metapneumovirus (hMPV) is a respiratory pathogen responsible for ARTIs in infants and children around the world. The epidemiology of hMPV in the northeast of Iran is poorly described. This study aims to evaluate the prevalence and molecular types of hMPV in children under the age of 12 along with clinical relevance in northeast Iran.Methods:
In this cross sectional study, from autumn 2015 to spring 2016 (nine months), 150 nasopharyngeal swabs were collected from children with ARTI and were evaluated for the presence of hMPV by reverse transcriptase-polymerase chain reaction (RT-PCR) technique; after that hMPV typing on positive samples was performed using PCR method.Results:
Among the 150 patients that were included the study, 31 (20.6%) were positive for the hMPV genome. We also found that 96.8% (n = 30 cases) of positive cases belonged to genotype A; whereas no B genotype was detected. One of our hMPV isolates was non typable (3.2%).Conclusions:
This study, for the first time, reports the existence of Human metapneumovirus virus in children with respiratory symptoms in northeastern Iran. We have also shown that the genotype A is the predominant circulating type among our patients.Keywords
Respiratory Tract Infection Human Metapneumovirus Child Iran
1. Background
Acute respiratory infection plays a major role in the hospitalization of children in the entire world. A wide range of bacterial and viral pathogens are responsible for acute respiratory tract infections (ARTIs), and newly emerging pathogens are reported every few years. Viruses are one of the most important causative agents of ARTIs, considered as the second leading cause of death and the primary cause of hospitalization in children (1-5). Due to worldwide incidence and also high prevalence of morbidity and mortality, respiratory viral infections, which easily spread, pose a serious public health problem (1-6). While Influenza virus, Para influenza virus, rhinovirus, and RSV are considered to be the most common respiratory viruses, the role of other new viruses such as hMPV in ARTI of children remains unknown; therefore, in this study we decided to determine the prevalence of hMPV in children with ARTI in northeastern Iran.
Van den Hoogen reported the isolation of a novel virus from nasopharyngeal aspirate samples taken from young children with respiratory tract infections in the Netherlands in 2001 (6). After that, human metapneumovirus has been reported as a respiratory pathogen repeatedly in children as well as in the elderly and the immune compromised host in different countries worldwide (1-6).
This virus is a new member of the family Paramyxoviridae, subfamily Pneumovirinae, and genus Metapneumovirus based on electron microscopy and molecular studies (7, 8). Like other members of the Paramyxoviridae family, hMPV is an enveloped, single-stranded, and negative-sense RNA virus. The genome is approximately 13.3 kb in length. Its genome encodes eight proteins (N, P, M, F, M2, SH, G, and L). According to the genetic property of the membrane glycoproteins F and G, hMPV has been divided into two main genotypes, A and B, that are in turn further subdivided into four subgroups A1, A2, B1, and B2 (9, 10).
hMPV is a causative agent of upper and lower respiratory tract infections in children and bronchiolitis in infants similar to that observed with RSV infection (11-16).
The symptoms related to hMPV range from a severe cough, fever, rhinorrhea, and influenza like symptoms to bronchiolitis, pneumonia, and wheezing attack (2, 17-19). Diagnosis of hMPV infections is now carried out by sensitive molecular techniques such as reverse transcription RT-PCR or, to a lesser extent, by isolation of virus in cell cultures and RT-PCR (20).
This study aims to determine the hMPV frequency in children with respiratory infection symptoms aged less than 12 years old, in Bojnurd and also determining the genotypes of hMPV in positive samples.
2. Methods
2.1. Patients and Samples
During October 2015 to June 2016, all children with acute respiratory tract infection (ARTI) symptoms referred to Imam Reza hospital, Bojnurd were included. Finally, nasopharyngeal swabs were collected from 150 children. Children less than six and more than 72 months as well as non compliant patient or patient’s legal guardian were excluded from this study. Samples were collected using a dacron swab that was preserved in a tube containing one mL PBS buffer. Samples were rapidly stored at -70°C until used.
2.2. RNA Extraction
All samples were thawed at room temperature, and viral genomic RNA was extracted from patients’ samples using Genetbio Prime preptm viral RNA/DNA extraction kit (Genetbio, Korea), according to the manufacturers’ provided protocol.
2.3. cDNA Synthesis and RT-PCR
cDNA synthesis was performed using QIAGEN Sensiscript® Reverse Transcription kit (Qiagen Germany), according to the manufacturer’s instructions.
Extracted RNA were used for detection of hMPV genome by the reverse transcriptase polymerase chain reaction with the common pair primer targeting a region of hMPV F gene described by Arabpour et al. (Gen Bank accession number AF371367) (21). The sequence of the forward and reverse primers used were, 5’-GCGCGTTCTGAGGACAGGTTGG-3’ and 5’- GCGCTCAAGCCGGATGGTTTTGG-3), respectively. PCR was performed using Genetbio Prime Taq premix 2X master mix (Korea). The PCR program comprised an initial denaturation at 95°C for 10 minutes, followed by 40 cycles of denaturation at 94°C for 1 minute, annealing at 57°C for 1 minute and extension at 72°C for 1 minute, and a final extension at 72°C for 10 minutes.
Amplified products were analyzed on 2% agarose gel by electrophoresis. Negative samples for hMPV genome were checked for the accurate performance of extraction and RT steps by carrying out a PCR reaction on RT products for Beta 2 microglobulin.
2.4. Molecular Typing
hMPV typing on positive samples was performed by PCR reaction using two sets of primers.
For type A, the sequence of the Forward primer was 5’-CATGCCCACTATAAAAGGTCAG-3’ and the sequence of the Reverse primer was 5’-CACCCCAGTCTTTCTTGAAA-3,’ amplifying a 170-bp segment of the L gene of hMPV as previously described conditions (21). For type B, the sequence of the Forward primer was 5’-TATGCCTACTATAAAAGGTCAA-3’; and the sequence of the Reverse Primer was 5’-CACCCCAGTCTTTCCTAAAG-3’, amplifying a 169 bp segment of the L gene of hMPV as previously described (21). Sequencing was done for one of the type A genomes.
2.5. Statistical Analysis
The statistical analysis was performed by SPSS 11.5 (Statistical packages for the Social Sciences, Chicago, IL). Chi-square and Mann-Whitney U tests were used for comparisons between positive and negative groups. A P value of less than 0.05 was considered statistically significant in all conclusions.
2.6. Ethical Considerations
The study was performed in accordance with Helsinki declaration on human research. Parental written informed consent, from all participants and children assent in possible cases, was obtained. The research proposal was reviewed and approved by local ethical committee in North Khorasan University of Medical Sciences (numbered IR.NKUMS.REC.1394.67).
3. Results
3.1. Demographic Data and Clinical Manifestations
A total of 150 outpatient children with respiratory tract infection were enrolled from October 2015 to Jun 2016. The ages of the children ranged from 6 months to 12 years old with a median ± IQR of 48 ± 42 months. No positive samples were found in children older than 6 years. In this study, 84 (56%) of the enrolled patients were female and 66 (44%) were male. There was no significant difference in two genders in terms of hMPV infection (P = 0.044). The median ± IQR age of hMPV positive and negative children was 30 ± 42 and 54 ± 48 months, respectively (Man-Whiney U Test P < 0.001).
hMPV was detected in 20.6% of samples (n = 31). The positive cases manifested by cough 100% (n = 31), wheezing 87% (n = 27), fever 64.5% (n = 20), sneezing 58% (n = 18), runny nose 48.38% (n = 15), bronchiolitis 48.38% (n = 15), rales 35.48% (n = 11), vomiting 35.48% (n = 11), otitis media 13% (n = 4), and asthma attack 9.6% (n = 3). There was no association between family history of asthma and positivity of h-MPV in patients with Chi-square test (P = 0.202). During the study period, hMPV infections occurred from January to May, with a peak in March (Figure 1).
Seasonal distribution of human metapneumovirus (hMPV) positive samples from October 2015 through Jun 2016 at Imam Reza hospital, Bojnurd

3.2. Molecular Diagnosis and Typing
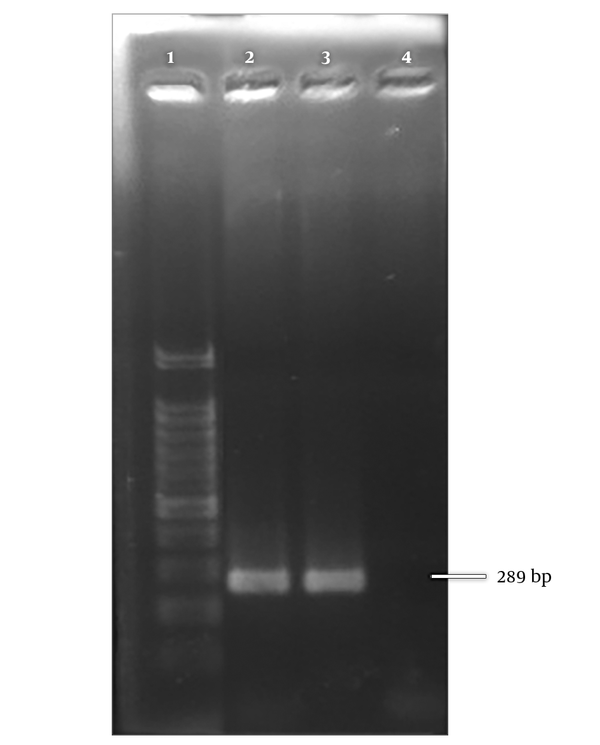
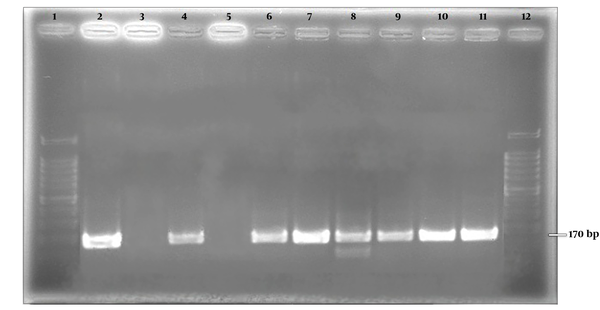
A 289 bp bond corresponding to amplicon size of Metapneumovirus was observed on gel electrophoresis (Figure 2). The positive samples were further subjected to more molecular investigation. Among positive samples, 30 (96.8%) belonged to genotype A and one (3.2%) of the positive cases was nontypable (Figure 3).
Positive control for hMPV. Lane 1; 100 bp ladder, lane 2; positive control, lane 3; positive clinical sample, lane 4; negative control.
Positive control for hMPV types A and B. Lane 1 , 12; 100 bp ladder, lane 2; positive control for type A, lane 3; negative control for type A, lane 4; Positive control for type B, lane 5; Negative control for type B, lanes 6 - 11; positive clinical samples for type A.
4. Discussion
Human metapneumovirus (hMPV), a newly pathogen of pediatric respiratory tract, is related to a wide range of respiratory symptoms from the common cold to pneumonia and bronchiolitis (1-6). In our attempt to identify this pathogen among children with respiratory tract infection symptoms, hMPV was detected in 31 out of 150 admitted cases (20.6%) indicating hMPV as a relatively common virus among these patients in Bojnurd, Iran.
A different frequency was reported by Arabpour et al. in Ahvaz-Iran where hMPV was positive in 54.4% of their samples in 2008 (21). Moattari et al. in 2010, reported that hMPV was detected in 16.6% of patients suffering from wheezing in Shiraz-Iran (3). Pancer et al. in 2014, reported that hMPV was positive in 8.7% of children with acute viral respiratory tract infection in Poland (22). In another study, Maffey et al. reported that only 10% of patients with acute wheezing were positive for hMPV; that was lower compared to our findings (23). These differences and similarities were also seen in other studies. Chan et al. in 2003, evaluated the prevalence of hMPV in severe acute respiratory syndrome (SARS). They reported 52.1% of hMPV colonization in SARS patients (3). Ebihara et al. in 2004, evaluated the hMPV Infection in Japanese Children. They reported hMPV in 36.8% of wheezy bronchitis, 26.3% of upper respiratory tract infection, 22.8% of bronchitis, and 14.0% of pneumonia patients (5). Peiris et al. reported different results of hMPV rate in various respiratory diseases. The virus was found in 36% of pneumonia, 23% of acute asthma, and 10% of acute bronchiolitis patients (24).
In concurrence with other studies (3, 21), the most common clinical symptom in hMPV positive children in this study were cough (100%) and wheezing (87%).
About 10% of our positive patients suffered from asthma. The association of hMPV and acute asthma symptoms have been reported by different studies (25-27).
In our study, prevalence of hMPV among male (45%) and female (55%) patients were almost similar. This result is in agreement with those of other studies (5, 21). Although in some studies, significant differences were detected, for example Moattari et al. reported that the prevalence of hMPV was three times greater in males than females (2).
In this study, sampling was performed during October to June; thus, our results could not be generalized to the whole year, though most studies have reported higher infection rates during winter and spring (2, 3, 7, 23, 25).
The prevalent genotype of our hMPV positive samples was A (96.8%). Various prevalent genotypes have been reported in different studies. Amer reported that in Saudi Arabia, in 2008, the majority of the hMPV strains belonged to subgroup B; whereas in 2009, both subgroups A and B were identified to be cocirculating at similar rates in another study (9). In opposition to our findings, Bayrakdar et al. reported that the most prevalent genotype in Turkey was B. According to sequences obtained, 71.7% belonged to B and 28.5% belonged to A (26). In concordance with the present study, Zhang et al. reported that the group A and B viruses accounted for 95.7% and 4.3% of infections, respectively (27). In addition, Aqrawal et al. reported 77% genotype A and 23% genotype B among hMPV positive samples (28). Some factors like geographical area, year, and season might affect the variety of prevalent genotype.
No positive samples were found in children older than 6 years. This finding could indicate that the acquisition of hMPV infection occurs early in life and leads to becoming immunologically resistant to the infection (29).
It should be noted that we did not rule out the presence of other respiratory viral pathogens in our hMPV positive children, therefore we can not conclude that hMPV was the sole causative agent of all ARTI in this study. To exact determination of hMPV role in ARTI of children should be further investigated in multi pathogen massive screening studies.
4.1. Conclusions
This is the first report of prevalence and genotypes of human metapneumovirus (hMPV) in children in northeastern Iran. The relatively noticeable frequency of hMPV (20.6%) indicates the clinical importance of this virus, especially type A, in our region. The rapid identification of these viral infections could help reduce the use of antibiotics and improve treatment and management strategies.
Acknowledgements
References
-
1.
Panda S, Mohakud NK, Suar M, Kumar S. Etiology, seasonality, and clinical characteristics of respiratory viruses in children with respiratory tract infections in Eastern India (Bhubaneswar, Odisha). J Med Virol. 2017;89(3):553-8. [PubMed ID: 27509268]. https://doi.org/10.1002/jmv.24661.
-
2.
Moattari A, Aleyasin S, Arabpour M, Sadeghi S. Prevalence of human Metapneumovirus (hMPV) in children with wheezing in Shiraz-Iran. Iran J Allergy Asthma Immunol. 2010;9(4):250-4. [PubMed ID: 21131706].
-
3.
Chan PK, Tam JS, Lam CW, Chan E, Wu A, Li CK, et al. Human metapneumovirus detection in patients with severe acute respiratory syndrome. Emerg Infect Dis. 2003;9(9):1058-63. [PubMed ID: 14519240]. [PubMed Central ID: PMC3016779]. https://doi.org/10.3201/eid0909.030304.
-
4.
Shatizadeh Malekshahi S, Mokhtari Azad T, Shahmahmoodi S, Yavarian J, Rezaei F, Naseri M. First report of respiratory syncytial virus and human metapneumovirus co-infection in a 2-year-old kawasaki patient in iran. Iran J Public Health. 2010;39(4):140-2. [PubMed ID: 23113048]. [PubMed Central ID: PMC3481691].
-
5.
Ebihara T, Endo R, Kikuta H, Ishiguro N, Ishiko H, Hara M, et al. Human metapneumovirus infection in Japanese children. J Clin Microbiol. 2004;42(1):126-32. [PubMed ID: 14715742]. [PubMed Central ID: PMC321731].
-
6.
van den Hoogen BG, Bestebroer TM, Osterhaus AD, Fouchier RA. Analysis of the genomic sequence of a human metapneumovirus. Virology. 2002;295(1):119-32. [PubMed ID: 12033771]. https://doi.org/10.1006/viro.2001.1355.
-
7.
Gaunt E, McWilliam-Leitch EC, Templeton K, Simmonds P. Incidence, molecular epidemiology and clinical presentations of human metapneumovirus; assessment of its importance as a diagnostic screening target. J Clin Virol. 2009;46(4):318-24. [PubMed ID: 19819755]. https://doi.org/10.1016/j.jcv.2009.09.016.
-
8.
Kahn JS. Human metapneumovirus: a newly emerging respiratory pathogen. Curr Opin Infect Dis. 2003;16(3):255-8. [PubMed ID: 12821817]. https://doi.org/10.1097/01.qco.0000073776.11390.bb.
-
9.
Amer HM. Molecular Epidemiology of Human Metapneumovirus in Riyadh Province, Saudi Arabia. J Mol Microbiol Biotechnol. 2016;26(6):414-21. [PubMed ID: 27694746]. https://doi.org/10.1159/000448374.
-
10.
Owor BE, Masankwa GN, Mwango LC, Njeru RW, Agoti CN, Nokes DJ. Human metapneumovirus epidemiological and evolutionary patterns in Coastal Kenya, 2007-11. BMC Infect Dis. 2016;16:301. [PubMed ID: 27316548]. [PubMed Central ID: PMC4912817]. https://doi.org/10.1186/s12879-016-1605-0.
-
11.
Yan XL, Li YN, Tang YJ, Xie ZP, Gao HC, Yang XM, et al. Clinical characteristics and viral load of respiratory syncytial virus and human metapneumovirus in children hospitaled for acute lower respiratory tract infection. J Med Virol. 2017;89(4):589-97. [PubMed ID: 27632796]. https://doi.org/10.1002/jmv.24687.
-
12.
Esposito S, Mastrolia MV. Metapneumovirus Infections and Respiratory Complications. Semin Respir Crit Care Med. 2016;37(4):512-21. [PubMed ID: 27486733]. https://doi.org/10.1055/s-0036-1584800.
-
13.
Taylor S, Lopez P, Weckx L, Borja-Tabora C, Ulloa-Gutierrez R, Lazcano-Ponce E, et al. Respiratory viruses and influenza-like illness: Epidemiology and outcomes in children aged 6 months to 10 years in a multi-country population sample. J Infect. 2017;74(1):29-41. [PubMed ID: 27667752]. https://doi.org/10.1016/j.jinf.2016.09.003.
-
14.
Bosis S, Esposito S, Niesters HG, Crovari P, Osterhaus AD, Principi N. Impact of human metapneumovirus in childhood: comparison with respiratory syncytial virus and influenza viruses. J Med Virol. 2005;75(1):101-4. [PubMed ID: 15543589]. https://doi.org/10.1002/jmv.20243.
-
15.
Bosis S, Esposito S, Niesters HG, Zuccotti GV, Marseglia G, Lanari M, et al. Role of respiratory pathogens in infants hospitalized for a first episode of wheezing and their impact on recurrences. Clin Microbiol Infect. 2008;14(7):677-84. [PubMed ID: 18558940]. https://doi.org/10.1111/j.1469-0691.2008.02016.x.
-
16.
Kuypers J, Wright N, Corey L, Morrow R. Detection and quantification of human metapneumovirus in pediatric specimens by real-time RT-PCR. J Clin Virol. 2005;33(4):299-305. [PubMed ID: 16036180]. https://doi.org/10.1016/j.jcv.2004.11.023.
-
17.
Williams JV, Crowe JE Jr, Enriquez R, Minton P, Peebles RJ, Hamilton RG, et al. Human metapneumovirus infection plays an etiologic role in acute asthma exacerbations requiring hospitalization in adults. J Infect Dis. 2005;192(7):1149-53. [PubMed ID: 16136455]. [PubMed Central ID: PMC1476781]. https://doi.org/10.1086/444392.
-
18.
Jartti T, Lehtinen P, Vuorinen T, Osterback R, van den Hoogen B, Osterhaus AD, et al. Respiratory picornaviruses and respiratory syncytial virus as causative agents of acute expiratory wheezing in children. Emerg Infect Dis. 2004;10(6):1095-101. [PubMed ID: 15207063]. [PubMed Central ID: PMC3323183]. https://doi.org/10.3201/eid1006.030629.
-
19.
Robinson JL, Lee BE, Bastien N, Li Y. Seasonality and clinical features of human metapneumovirus infection in children in Northern Alberta. J Med Virol. 2005;76(1):98-105. [PubMed ID: 15778961]. https://doi.org/10.1002/jmv.20329.
-
20.
Percivalle E, Sarasini A, Visai L, Revello MG, Gerna G. Rapid detection of human metapneumovirus strains in nasopharyngeal aspirates and shell vial cultures by monoclonal antibodies. J Clin Microbiol. 2005;43(7):3443-6. [PubMed ID: 16000473]. [PubMed Central ID: PMC1169090]. https://doi.org/10.1128/JCM.43.7.3443-3446.2005.
-
21.
Arabpour M, Samarbafzadeh AR, Makvandi M, Shamsizadeh A, Percivalle E, Englud J, et al. The highest prevalence of human metapneumovirus in Ahwaz children accompanied by acute respiratory infections. Indian J Med Microbiol. 2008;26(2):123-6. [PubMed ID: 18445946].
-
22.
Pancer KW, Gut W, Abramczuk E, Lipka B, Litwinska B. Non-influenza viruses in acute respiratory infections among young children. High prevalence of HMPV during the H1N1V.2009 pandemic in Poland. Przegl Epidemiol. 2014;68(4):627-32. [PubMed ID: 25848781].
-
23.
Maffey AF, Venialgo CM, Barrero PR, Fuse VA, Marques Mde L, Saia M, et al. [New respiratory viruses in children 2 months to 3 years old with recurrent wheeze]. Arch Argent Pediatr. 2008;106(4):302-9. [PubMed ID: 18766276]. https://doi.org/10.1590/S0325-00752008000400005.
-
24.
Peiris JS, Tang WH, Chan KH, Khong PL, Guan Y, Lau YL, et al. Children with respiratory disease associated with metapneumovirus in Hong Kong. Emerg Infect Dis. 2003;9(6):628-33. [PubMed ID: 12781000]. [PubMed Central ID: PMC3000155]. https://doi.org/10.3201/eid0906.030009.
-
25.
Shatizadeh S, Yavarian J, Rezaie F, Mahmoodi M, Naseri M, Mokhtari Azad T. Epidemiological and clinical evaluation of children with respiratory virus infections. Med J Islam Repub Iran. 2014;28:102. [PubMed ID: 25664303]. [PubMed Central ID: PMC4301222].
-
26.
Bayrakdar F, Altas AB, Korukluoglu G. [Phylogenetic variability of human metapneumovirus strains circulating in Turkey during two consecutive epidemic seasons]. Mikrobiyol Bul. 2016;50(1):63-72. [PubMed ID: 27058330].
-
27.
Zhang C, Du LN, Zhang ZY, Qin X, Yang X, Liu P, et al. Detection and genetic diversity of human metapneumovirus in hospitalized children with acute respiratory infections in Southwest China. J Clin Microbiol. 2012;50(8):2714-9. [PubMed ID: 22692746]. [PubMed Central ID: PMC3421497]. https://doi.org/10.1128/JCM.00809-12.
-
28.
Agrawal AS, Roy T, Ghosh S, Chawla-Sarkar M. Genetic variability of attachment (G) and Fusion (F) protein genes of human metapneumovirus strains circulating during 2006-2009 in Kolkata, Eastern India. Virol J. 2011;8:67. [PubMed ID: 21314961]. [PubMed Central ID: PMC3045894]. https://doi.org/10.1186/1743-422X-8-67.
-
29.
Berry M, Gamieldien J, Fielding BC. Identification of new respiratory viruses in the new millennium. Viruses. 2015;7(3):996-1019.
