Abstract
Background:
Cronobacter is a genus within the family Enterobacteriaceae and was previously known as Enterobacter sakazakii. Cronobacter sakazakii is an opportunistic ubiquitous bacterium and is identified throughout the world as an emerging food-borne pathogen. This pathogen causes meningitis, necrotizing enterocolitis (NEC), and sepsis (septicemia) in patients hospitalized in neonatal intensive care units and has a high mortality and morbidity rate. Contamination of powdered infant milk formula occurs more easily because it is a nonsterilized product.Objectives:
The aims of the present study were as follows: Isolation of Cronobacter sakazakii from powdered infant formula milk (PIF), confirmation of isolates by biochemical tests and polymerase chain reaction (PCR) molecular method, and characterization of an antibiotic susceptibility profile.Methods:
A cross-sectional study was performed on 125 powdered infant formula milk (PIF) samples purchased from hospital drug stores between June 2014 and March 2015. Cronobacter sakazakii was recovered according to FDA protocol. For the final confirmation of isolates, different biochemical tests embedded in the API-20E kit system and manual biochemical tests were used according to the directions of the manufacturer. Antibiotic susceptibility testing was performed on Mueller-Hinton agar by disk diffusion method according to CLSI recommendations. For molecular confirmation of isolates confirmed by biochemical tests, PCR was performed on 16S rRNA gene specific for Cronobacter sakazakii.Results:
Out of the 125 PIF samples investigated, nine (7.2%) of the samples were positive for Cronobacter sakazakii. All C. sakazakii strains isolated from PIF samples were uniformly susceptible to ticarcillin, chloramphenicol, levofloxacin, ciprofloxacin, moxifloxacin, cefepime, piperacillin-tazobactam, and colistin. All isolates were confirmed by PCR.Conclusions:
Results showed that PIF that is being consumed in Iran is contaminated with this pathogen and can cause disease in infants; especially those hospitalized in neonatal intensive care units (NICUs) and fed PIF. The number of reported cases of Cronobacter infections is very low, but nevertheless there has been a slight increase recently. While the reported cases worldwide are few, it needs to be noted that the number of infections may be underestimated since not all clinical analysis laboratories carry out research on other bacterial pathogens, and not all countries have a system for reporting diseases.Keywords
Infant Infant Formula Foodborne Disease Polymerase Chain Reaction Cronobacter sakazakii
1. Background
Cronobacter is a genus within the family Enterobacteriaceae and was previously known as Enterobacter sakazakii. Cronobacter are gram-negative, motile by peritrichous flagella, rod-shaped, and non-spore-forming bacteria. It is closely related to the genera Enterobacter and Citrobacter. Cronobacter spp. have been frequently isolated from the environment, plant material (wheat, rice, herbs, and spices), and various food products, including powdered infant formula (PIF) (1, 2). Cronobacter sakazakii is identified throughout the world as an emerging food-borne pathogen. It is distributed in the environment, vegetables, fermented beverages, PIF, cereal foods, fruits, and plant materials (3, 4). In particular, contamination of PIF occurs more easily because it is a nonsterilized product (5).
Our current knowledge of the virulence and epidemiology of the Cronobacter organism is limited. However, because neonates (especially infants hospitalized in neonatal intensive care units) are frequently fed reconstituted PIF, this product has been the focus of attention for decreasing infection risk to neonates because the number of exposure routes is restricted (1, 2). Cronobacter sakazakii is an opportunistic pathogen. Infections caused by Cronobacter occur across all age groups and most infections although less severe are in the adult population. However, neonates particularly those of low birth weight (LBW), preterm neonates, and immunocompromised infants are the major recognized group at particular risk because the bacterium causes meningitis, necrotizing enterocolitis (NEC), and sepsis (septicemia) in patients hospitalized in neonatal intensive care units (NICUs) and has high mortality and morbidity rate (1-3, 6, 7). About 10% - 80% mortality rates have been reported, and surviving patients often suffer from neurological problems and sequel. Cronobacter infection outbreaks in NICUs related to contaminate PIF have been described (7, 8). Cronobacter sakazakii is considered an opportunistic pathogen that has been implicated in severe forms of meningitis (leading to brain abscess, ventriculitis, infarction, and cyst formation) and necrotizing enterocolitis, especially in neonates, with a mortality rate varying from 40% - 80% (9, 10).
Cronobacter sakazakii is a ubiquitous foodborne microorganism. The source of this organism and route of transmission to humans is not always clear, but PIF has been implicated as the source of this pathogenic bacterium in several patients. The source of contamination in PIF is thought to include a broad range of dry blended raw material (e.g., rice, soybean) together with environmental sources related to the production environment (5, 6, 11-13). The international commission for microbiological specifications for foods (ICMSF) has classified Cronobacter sakazakii as a severe hazard for restricted populations (including infants), including those afflicted with substantial chronic sequelae or life threatening diseases or those who are infected for a long duration (14). As a result, C. sakazakii has been included among the most common bacterial pathogens, along with A Clostridium botulinum and Listeria monocytogenes (15). Studies reported that C. sakazakii, in a similar route as Salmonella, is increasingly causing the most concern among foodborne bacterium. It has been noted that the attendance of this bacterium represents a serious risk if the conditions following the reconstitution of PIF allow its replication (4, 16). The main goal of the present research was to detect Cronobacter sakazakii strains in powdered milk infant formula that is being consumed in intensive care units in hospitals by phenotypic and molecular methods.
2. Objectives
The aims of present study were to isolate Cronobacter sakazakii from powdered infant formula milk (PIF), confirm isolates by biochemical test and PCR molecular method, and characterize the antibiotic susceptibility profile.
3. Methods
3.1. Sampling
A cross-sectional study was carried out on 125 powdered infant formula milk (PIF) samples purchased from hospital drug stores between Jun 2014 and March 2015.
3.2. Isolation and Identification
3.2.1. Cronobacter sakazakii
For isolation of C. sakazakii from samples, the surfaces of PIF cans were sterilized with 70% ethanol and were aseptically opened in a laminar flow cabinet. Samples were taken from each product under aseptic conditions. Cronobactersakazakii was recovered according to FDA protocol (17, 18). We prepared three Erlenmeyer flasks each of sterile distilled water (pre-warmed to 45°C) at 9, 90 and 900 mL containing 1, 10 and 100 g of PIF, respectively. After the weighed PIF was completely mixed and dissolved in distilled water, it was incubated at 35 ± 2°C for 18 to 24 hours. After incubation, 10 ml of each sample was added to 90 mL of EE (Enterobacteriaceae enrichment) broth medium (Merck Co., Germany) and placed at 35 ± 2°C for 18 to 24 hours. Following the incubation, a loopful of the enrichment culture was streaked onto duplicate VRBGA (violet red bile glucose agar) medium (Merck Co., Germany) plates and incubation was performed for 18 - 24 hours at 35 ± 2°C. Four presumptive colonies were picked from each VRBGA plate, and pure culture was performed on MacConkey agar and TSA (tryptic soy agar) (Merck Co., Germany). The isolated colonies that produce yellow pigment on TSA medium in room temperature (25°C) were identified. For the final confirmation of isolates, manual biochemical tests and biochemical tests embedded in API-20E biochemical kit system (Bio-Merieux, France) were used according to the recommendations of the manufacturer. Finally, the purified isolates were saved in TSB (tryptic soy broth) supplemented with 20% glycerol (Merck Co., Germany) at -20°C (17, 18).
3.2.2. Antimicrobial Susceptibility Testing of Isolates
Antibiotic susceptibility testing was done using Kirby-Bauer disk diffusion method on Mueller Hinton agar (Merck Co., Germany) according to CLSI guidelines (19). Antimicrobial agents (MAST Co., UK) used in this study were ampicillin (AP, 10μg), piperacillin (PRL, 100μg), piperacillin-tazobactam (PTZ, 100/10μg), mezlocillin (MZ, 75μg), carbenicillin (PY, 100μg), ticarcillin (TC, 75μg), ceftazidime (CAZ, 30μg), cefotaxime (CTX, 30μg), ceftriaxone (CRO, 30μg), cefepime (CPM, 30μg), imipenem (IMI, 10μg), meropenem (MEM, 10μg), aztreonam (AZT, 30μg), chloramphenicol (C, 30μg), amikacin (AK, 30μg), streptomycin (S, 10μg), tobramycin (TN, 10μg), gentamicin (GM, 10μg), tetracycline (T, 30μg), minocycline (MN, 30μg), tigecycline (TG, 15μg), ciprofloxacin (CIP, 5μg), levofloxacin (LEV, 5μg), moxifloxacin (MFX, 5μg), colistin (CO, 25μg), trimethoprim-sulfamethoxazole (SXT, 1.25/23.75μg) and nalidixic acid (NA, 30μg). Escherichia coli ATCC 25922 was applied for quality control (QC) of antimicrobial susceptibility tests.
3.2.3. PCR Assay
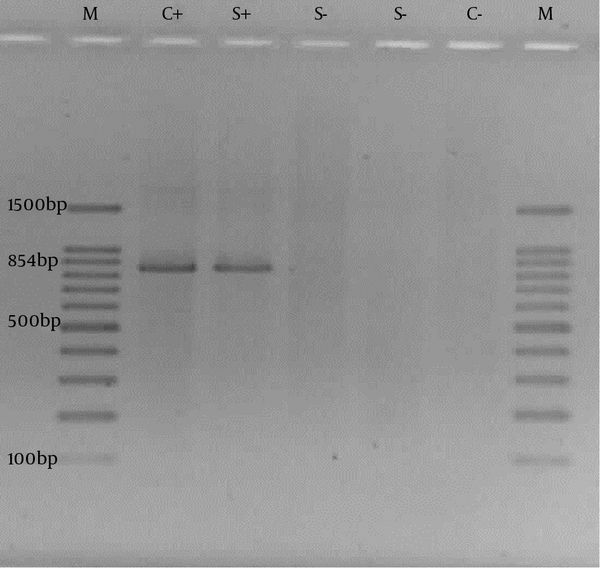
For confirmation of the biochemical species identification, putative Cronobacter sakazakii isolates were further analyzed using the molecular method. PCR was performed on 16S rRNA gene specific for Cronobacter sakazakii (20). Genomic DNA was extracted from overnight cultures. DNA templates for PCR experiments were prepared by simply boiling a dense bacterial suspension in ddH2O and performing 10 minutes of centrifugation at 13000 rpm (21). PCR reactions were set in 25μL final volumes consisting of 5 μL of the bacterial genome DNA, 1 × buffer, 1.5 U Taq polymerase (Fermentas, Lithuania), 1.5 mM MgCl2, 0.8 mM dNTPs and 20 pmol primers each. Primers, Esak2-5′-CCC GCA TCT CTG CAG GAT TCT C-3′ and Esak3 -5′-CTA ATA CCG CAT AAC GTC TAC G-3′ were used (59°C annealing temperature) in PCR to amplify an 854 bp fragment to 16S rRNA gene (20). The PCR was carried out with initiation at 94°C for 5 minutes, then 30 cycles of denaturation at 94°C for 30 seconds, annealing at 59°C for 30 seconds and extension at 72°C for 2 minutes, followed by a terminal extension step of 72°C for 5 minutes. When PCR amplification was completed, the PCR products were analyzed by 1% (w/v) agarose gel electrophoresis in 1X TAE buffer (40 mM Tris-HCl, 1.18 mL acetic acid, 2 mM EDTA, pH 8.0). The gel was stained with ethidium bromide (EtBr) for 25 minutes in 1X TAE buffer containing 0.5 μg/mL EtBr and visualized using UV transilluminator system (Gel Doc XR). Cronobacter sakazakii ATCC 29544 was used as positive control in molecular assay.
4. Results
Out of the 125 PIF samples studied, 9 (7.2%) of the samples were positive for Cronobacter sakazakii. The frequency of other Enterobacter species family members isolated from PIF samples are listed in Table 1. All C. sakazakii strains isolated from PIF samples were uniformly susceptible to ticarcillin, chloramphenicol, levofloxacin, ciprofloxacin, moxifloxacin, cefepime, piperacillin-tazobactam and colistin. One isolate was resistant to meropenem and imipenem. Cefotaxime and ceftazidime were less effective against these isolates.
Frequency and Susceptibility Patterns of Enterobacter Species Isolated From PIF Samples
| E. cowanii (n = 4) | E. aminogenus (n = 2) | E. asburiae (n = 2) | E. sakazakii (n = 9) | E. aglumerans (n = 8) | E. colaceae (n = 2) | |
|---|---|---|---|---|---|---|
| Amoxicillin | 1 (25) | 0 | 1 (50) | 8 (88.8) | 2 (25) | 0 |
| Ampicillin | 1 (25) | 0 | 1 (50) | 8 (88.8) | 1 (12.5) | 0 |
| Aztreonam | 3 (75) | 2 (100) | 2 (100) | 8 (88.8) | 6 (75) | 1 (50) |
| Cefotaxime | 1 (25) | 2 (100) | 2 (100) | 2 (22.2) | 3 (37.5) | 0 |
| Ceftazidime | 4 (100) | 2 (100) | 2 (100) | 9 (100) | 6 (75) | 2 (100) |
| Imipenem | 3 (75) | 1 (50) | 2 (100) | 9 (100) | 6 (75) | 2 (100) |
| Meropenem | 3 (75) | 2 (100) | 2 (100) | 8 (88.8) | 5 (62.5) | 2 (100) |
| Mezlocillin | 2 (50) | - | 1 (50) | 3 (33.3) | 3 (37.5) | - |
| Ticarcillin | 3 (75) | 1 (50) | 1 (50) | 9 (100) | 4 (50) | 2 (100) |
| Moxifloxacin | 4 (100) | 2 (100) | 2 (100) | 9 (100) | 4 (50) | 2 (100) |
| Nalidixic Acid | 3 (75) | 2 (100) | 2 (100) | 8 (88.8) | 5 (62.5) | 2 (100) |
| Streptomycin | 3 (75) | 2 (100) | 2 (100) | 3 (33.3) | 6 (75) | 0 |
| Gentamicin | 4 (100) | 2 (100) | 1 (50) | 6 (66.6) | 6 (75) | 1 (50) |
| Amikacin | 3 (75) | 2 (100) | 2 (100) | 6 (66.6) | 5 (62.5) | 1 (50) |
| Tetracycline | 3 (75) | 2 (100) | 2 (100) | 8 (88.8) | 7 (87.5) | 2 (100) |
| Chloramphenicol | 4 (100) | 2 (100) | 0 | 9 (100) | 8 (100) | 2 (100) |
| Levofloxacin | 4 (100) | 2 (100) | 2 (100) | 9 (100) | 8 (100) | 2 (100) |
| Ciprofloxacin | 4 (100) | 2 (100) | 2 (100) | 9 (100) | 7 (87.5) | 2 (100) |
| Cefepime | 4 (100) | 2 (100) | 2 (100) | 9 (100) | 8 (100) | 2 (100) |
| Minocycline | 3 (75) | 2 (100) | 1 (50) | 1 (11.1) | 8 (100) | 2 (100) |
| Piperacillin | 4 (100) | 1 (50) | 1 (50) | 7 (77.7) | 5 (62.5) | 2 (100) |
| Piperacillin-tazobactam | 4 (100) | 2 (100) | 2 (100) | 9 (100) | 5 (62.5) | 2 (100) |
| Carbenicillin | 1 (25) | 1 (25) | - | 6 (66.6) | 1 (12.5) | 1 (50) |
| Tobramycin | 3 (75) | 2 (100) | 1 (50) | 4 (44.4) | 7 (87.5) | 2 (100) |
| Cotrimoxazole | 2 (50) | 1 (50) | 2 (100) | 3 (33.3) | 4 (50) | 1 (50) |
| Tigecycline | 1 (25) | 2 (100) | 1 (50) | 4 (44.4) | 8 (100) | 2 (100) |
| Colistin | 4 (100) | 2 (100) | 9 (100) | 8 (100) | 2 (100) |
Variable antibiotic susceptibility pattern was seen to the other antimicrobial agents surveyed, and 88.8% of isolates were sensitive to amoxicillin, ampicillin, aztreonam, nalidixic acid and tetracycline, while 33.3% were susceptible to mezlocillin, streptomycin and cotrimoxazole. The response to amikacin, carbenicillin, tobramycin and tigecycline was 66.6%, 66.6%, 44.4%, and 44.4%, respectively.
PCR on 16S rRNA gene was performed on isolates that were confirmed by biochemical tests as C. sakazakii. All isolates were confirmed by PCR. The agarose gel electrophoresis results are shown in Figure 1.
PCR Results on 16S rRNA Gene of Cronobacter sakazakii.
5. Discussion
A large part of Cronobacter sakazakii infections occur in hospitals, especially in the NICU. Keeping reconstituted PIF at room temperature (home or hospital) for long periods of time, in fact, increases the risk of C. sakazakii infection. That is why improving training and hygiene in hospitals (especially NICU wards) is very important in preventing C.sakazakii outbreaks among hospitalized neonates (22). Due to the seriousness of pathologies associated with C. sakazakii, the ICMSF defined it a “serious danger for a segment of the population, with chronic effects which may be long-lasting, capable of constituting a threat for life” (23).
In the past decade, different data has been reported on the prevalence of various Cronobacter isolates in PIF based on conventional and molecular methods. Very little information is available, however, on the prevalence of Cronobacter in Iran. Our results for meropenem resistance among C.sakazaii (11.2%), which is thought to be the result of carbapenemase production, are not in agreement with some reports from other countries like the US, which showed the resistance to meropenem to be 4%. In the present study, all (100%) of C. sakazaki isolates were susceptible to ciprofloxacin and levofloxacin, which is much higher than the Kilonzo-Nthenge report from Tennessee, United States (42.9%) in 2012. In our study, 88.8% of the isolates were sensitive to tetracycline and nalidixic acid, but in other studies sensitivity was reported at 33.4% and 52.4%, respectively (24). As such an antibiotic resistance can easily spread within hospital wards and cause protracted outbreaks with high mortality and morbidity rates, strict infection control protocols are recommended (24). In the present study, 11.2% of isolates were resistant to ampicillin; similarly, previous studies performed by Kim et al. (2008) (25) reported 31.6% of isolated strains were resistant to ampicillin.
To our knowledge, this is the first report that has applied the phenotypic and molecular methods to study Cronobacter sakazakii in Iran. In the study of Ahmadi and associates (26) on the molecular detection of C. sakazakii in blood specimens of hospitalized neonates suspected to have sepsis in Ahvaz, this bacterium is not identified. A microbiological analysis performed by Reich et al. (27) in a PIF processing environment revealed that the environment was correlated with Cronobacter contamination in the final products, suggesting that the processing environment may be a contamination source. Moreover, contamination of PIF with Cronobacter after processing is also important and should not be overlooked. Infants, especially those hospitalized in NICUs and fed with PIF, are the group of population most seriously affected by Cronobacter. The Codex Committee on Food Hygiene of FAO/WHO is considering the creation of a risk pattern for C. sakazakii in PIF (28).
Cronobacter sakazakii may cause infections in all age groups (particularly in infants of 0 to 12 months). Premature infants born after less than 36 weeks, underweight infants, infants with immunodeficiency, infants whose mothers are HIV-positive and infants hospitalized in intensive care units are more at risk of infection. The reason is that they are usually fed with PIF, which is the most common route of transmission of the bacterium (22, 29). Different foodborne disease-causing opportunistic bacteria are included in the Enterobacteriaceae family, and the point to be noted is that immunosuppressed persons are prime candidates for infections caused by these opportunistic pathogens (30-34).
Some of the vehicles of transmission and the infectious dose of Cronobacter sakazakii are unknown. The number of reported cases of Cronobacter infections is very low, but nevertheless it has slightly increased recently. While the reported cases worldwide are less frequent, it needs to be noted that the number of infections may be underestimated since not all clinical analysis laboratories carry out research on other bacterial pathogens, and not all countries (such as Iran) have a system for reporting diseases.
Since Cronobacter sakazakii does not withstand the temperatures at which milk is pasteurized but is easily found post-pasteurization, manufacturer environment, as well as preparation and handling before consumption, represents critical points. This is the reason why important steps towards preventive action are needed to eliminate or avoid risks of PIF contamination and replication of the C. sakazakii. It is worth pointing out that breastfeeding should always be supported and encouraged since a mother’s milk constitutes the preferred food for newborn infants, especially in their early months. When this is not possible, a mother should be well informed and educated on the importance of hygiene while handling, preparing and storing PIF. Recommendations also underline the importance of using ready-to-feed PIF, complying with the rules for aseptic preparation and refrigeration at 2 to 3°C of reconstituted PIF for a span of time shorter than 4 hours.
The point to be noted here is that detecting C. sakazaki in PIF is very important, and results showed that PIF that is being consumed in Iran is contaminated with this pathogen and can cause disease in hospitalized patients. Thus, additional studies on this foodborne pathogen are necessary.
References
-
1.
Joseph S, Forsythe SJ. Predominance of Cronobacter sakazakii sequence type 4 in neonatal infections. Emerg Infect Dis. 2011;17(9):1713-5. [PubMed ID: 21888801]. https://doi.org/10.3201/eid1709.110260.
-
2.
Chap J, Jackson P, Siqueira R, Gaspar N, Quintas C, Park J, et al. International survey of Cronobacter sakazakii and other Cronobacter spp. in follow up formulas and infant foods. Int J Food Microbiol. 2009;136(2):185-8. [PubMed ID: 19729216]. https://doi.org/10.1016/j.ijfoodmicro.2009.08.005.
-
3.
Drudy D, Mullane NR, Quinn T, Wall PG, Fanning S. Enterobacter sakazakii: an emerging pathogen in powdered infant formula. Clin Infect Dis. 2006;42(7):996-1002. [PubMed ID: 16511766]. https://doi.org/10.1086/501019.
-
4.
Fiore A, Casale M, Aureli P. Enterobacter sakazakii: epidemiology, clinical presentation, prevention and control. Ann Ist Super Sanita. 2008;44(3):275-80. [PubMed ID: 18946167].
-
5.
Beuchat LR, Kim H, Gurtler JB, Lin LC, Ryu JH, Richards GM. Cronobacter sakazakii in foods and factors affecting its survival, growth, and inactivation. Int J Food Microbiol. 2009;136(2):204-13. [PubMed ID: 19346021]. https://doi.org/10.1016/j.ijfoodmicro.2009.02.029.
-
6.
Baumgartner A, Grand M, Liniger M, Iversen C. Detection and frequency of Cronobacter spp. (Enterobacter sakazakii) in different categories of ready-to-eat foods other than infant formula. Int J Food Microbiol. 2009;136(2):189-92. [PubMed ID: 19419789]. https://doi.org/10.1016/j.ijfoodmicro.2009.04.009.
-
7.
Caubilla-Barron J, Hurrell E, Townsend S, Cheetham P, Loc-Carrillo C, Fayet O, et al. Genotypic and phenotypic analysis of Enterobacter sakazakii strains from an outbreak resulting in fatalities in a neonatal intensive care unit in France. J Clin Microbiol. 2007;45(12):3979-85. [PubMed ID: 17928419]. https://doi.org/10.1128/JCM.01075-07.
-
8.
Dumen E. Cronobacter sakazakii (Enterobacter sakazakii). Only An Infant Problem? Kafkas Univ Vet Fak Derg. 2010;16:171-8.
-
9.
Muytjens HL, Roelofs-Willemse H, Jaspar GH. Quality of powdered substitutes for breast milk with regard to members of the family Enterobacteriaceae. J Clin Microbiol. 1988;26(4):743-6. [PubMed ID: 3284901].
-
10.
Shaker R, Osaili T, Al-Omary W. Isolation of Enterobacter sakazakii and other Enterobacter sp. from food and food production environments. Food Control. 2007;18:1241-5. https://doi.org/10.1016/j.foodcont.2006.07.020.
-
11.
Li R, Fei P, Man CX, Lou BB, Niu JT, Feng J, et al. Tea polyphenols inactivate Cronobacter sakazakii isolated from powdered infant formula. J Dairy Sci. 2016;99(2):1019-28. [PubMed ID: 26627860]. https://doi.org/10.3168/jds.2015-10039.
-
12.
Iversen C, Mullane N, McCardell B, Tall BD, Lehner A, Fanning S, et al. Cronobacter gen. nov., a new genus to accommodate the biogroups of Enterobacter sakazakii, and proposal of Cronobacter sakazakii gen. nov., comb. nov., Cronobacter malonaticus sp. nov., Cronobacter turicensis sp. nov., Cronobacter muytjensii sp. nov., Cronobacter dublinensis sp. nov., Cronobacter genomospecies 1, and of three subspecies, Cronobacter dublinensis subsp. dublinensis subsp. nov., Cronobacter dublinensis subsp. lausannensis subsp. nov. and Cronobacter dublinensis subsp. lactaridi subsp. nov. Int J Syst Evol Microbiol. 2008;58(Pt 6):1442-7. [PubMed ID: 18523192]. https://doi.org/10.1099/ijs.0.65577-0.
-
13.
Lee HA, Hong S, Park H, Kim H, Kim O. Cronobacter sakazakii Infection Induced Fatal Clinical Sequels Including Meningitis in Neonatal ICR Mice. Lab Anim Res. 2011;27(1):59-62. [PubMed ID: 21826162]. https://doi.org/10.5625/lar.2011.27.1.59.
-
14.
Feeney A, Kropp KA, O'Connor R, Sleator RD. Cronobacter sakazakii: stress survival and virulence potential in an opportunistic foodborne pathogen. Gut Microbes. 2014;5(6):711-8. [PubMed ID: 25562731]. https://doi.org/10.4161/19490976.2014.983774.
-
15.
Iversen C, Forsythe S. Risk profile of Enterobacter sakazakii, an emergent pathogen associated with infant milk formula. Trends Food Sci Technol. 2003;14:443-54. https://doi.org/10.1016/S0924-2244(03)00155-9.
-
16.
Kalyantanda G, Shumyak L, Archibald LK. Cronobacter Species Contamination of Powdered Infant Formula and the Implications for Neonatal Health. Front Pediatr. 2015;3:56. [PubMed ID: 26191519]. https://doi.org/10.3389/fped.2015.00056.
-
17.
International Special dietary Foods Industries. Background information on Enterobacter sakazakii. 2004, [updated 08/01/2007]. Available from: http://www.awex.be/NR/rdonlyres/A3A18BBA-F13C-4D36-97B1-26519CB2E9EA/4055/Enterobacter.
-
18.
Mardaneh J, Soltan-Dallal MM. Isolation and Identification of E. cowanii from Powdered Infant Formula in NICU and Determination of Antimicrobial Susceptibility of Isolates. Iran J Pediatr. 2014;24(3):261-6. [PubMed ID: 25562018].
-
19.
Poorabbas B, Mardaneh J, Rezaei Z, Kalani M, Pouladfar G, Alami MH, et al. Nosocomial Infections: Multicenter surveillance of antimicrobial resistance profile of Staphylococcus aureus and Gram negative rods isolated from blood and other sterile body fluids in Iran. Iran J Microbiol. 2015;7(3):127-35. [PubMed ID: 26668699].
-
20.
Keyser M, Witthuhn RC, Ronquest LC, Britz TJ. Treatment of winery effluent with upflow anaerobic sludge blanket (UASB)--granular sludges enriched with Enterobacter sakazakii. Biotechnol Lett. 2003;25(22):1893-8. [PubMed ID: 14719823].
-
21.
Mitov I, Strateva T, Markova B. Prevalence of virulence genes among bulgarian nosocomial and cystic fibrosis isolates of pseudomonas aeruginosa. Braz J Microbiol. 2010;41(3):588-95. [PubMed ID: 24031533]. https://doi.org/10.1590/S1517-83822010000300008.
-
22.
Masood N, Moore K, Farbos A, Paszkiewicz K, Dickins B, McNally A, et al. Genomic dissection of the 1994 Cronobacter sakazakii outbreak in a French neonatal intensive care unit. BMC Genomics. 2015;16:750. [PubMed ID: 26438044]. https://doi.org/10.1186/s12864-015-1961-y.
-
23.
Microbiological testing in food safety management. New York: Kluwer cademic/Plenum Publishers; 2002.
-
24.
Kilonzo-Nthenge A, Rotich E, Godwin S, Nahashon S, Chen F. Prevalence and antimicrobial resistance of Cronobacter sakazakii isolated from domestic kitchens in middle Tennessee, United States. J Food Prot. 2012;75(8):1512-7. [PubMed ID: 22856579]. https://doi.org/10.4315/0362-028X.JFP-11-442.
-
25.
Kim K, Jang SS, Kim SK, Park JH, Heu S, Ryu S. Prevalence and genetic diversity of Enterobacter sakazakii in ingredients of infant foods. Int J Food Microbiol. 2008;122(1-2):196-203. [PubMed ID: 18177966]. https://doi.org/10.1016/j.ijfoodmicro.2007.11.072.
-
26.
Ahmadi K, Farajzadeh Sheikh A, Mardaneh J, Modarresi F, Shoja S. Detection of Enterobacter sakazakii in neonatal sepsis by PCR on 16S ribosomal RNA. Iranian South Med J. 2014;17(3):272-9.
-
27.
Reich F, Konig R, von Wiese W, Klein G. Prevalence of Cronobacter spp. in a powdered infant formula processing environment. Int J Food Microbiol. 2010;140(2-3):214-7. [PubMed ID: 20409601]. https://doi.org/10.1016/j.ijfoodmicro.2010.03.031.
-
28.
Masood N, Moore K, Farbos A, Hariri S, Paszkiewicz K, Dickins B, et al. Draft Genome Sequences of Three Newly Identified Species in the Genus Cronobacter, C. helveticus LMG23732T, C. pulveris LMG24059, and C. zurichensis LMG23730T. Genome Announc. 2013;1(5). [PubMed ID: 24072872]. https://doi.org/10.1128/genomeA.00783-13.
-
29.
Farber JM. Enterobacter sakazakii--new foods for thought? Lancet. 2004;363(9402):5-6. [PubMed ID: 14723984]. https://doi.org/10.1016/S0140-6736(03)15244-0.
-
30.
Mardaneh J, Soltan Dallal MM, Taheripoor M, Rajabi Z. Isolation, Identification and Antimicrobial Susceptibility Pattern of Tatumella ptyseos Strains Isolated From Powdered Infant Formula Milk Consumed in Neonatal Intensive Care Unit: First Report From Iran. Jundishapur J Microbiol. 2014;7(6):10608. [PubMed ID: 25371802]. https://doi.org/10.5812/jjm.10608.
-
31.
Mardaneh J, Dallal MM. Isolation, identification and antimicrobial susceptibility of Pantoea (Enterobacter) agglomerans isolated from consumed powdered infant formula milk (PIF) in NICU ward: First report from Iran. Iran J Microbiol. 2013;5(3):263-7. [PubMed ID: 24475334].
-
32.
Shaghaghian S, Pourabbas B, Alborzi A, Askarian M, Mardaneh J. Vancomycin-Resistant Entrococci colonization in chronic hemodialysis patients and its risk factors in southern Iran (2005-2006). Iran Red Crescent Med J. 2012;14(10):686-91. [PubMed ID: 23285424].
-
33.
Anvarinejad M, Pouladfar GR, Pourabbas B, Amin Shahidi M, Rafaatpour N, Dehyadegari MA, et al. Detection of Salmonella spp. with the BACTEC 9240 Automated Blood Culture System in 2008 - 2014 in Southern Iran (Shiraz): Biogrouping, MIC, and Antimicrobial Susceptibility Profiles of Isolates. Jundishapur J Microbiol. 2016;9(4):26505. [PubMed ID: 27284396]. https://doi.org/10.5812/jjm.26505.
-
34.
Mardaneh J, Soltan Dallal MM. Isolation and Identification Enterobacter asburiae from Consumed Powdered Infant Formula Milk (PIF) in the Neonatal Intensive Care Unit (NICU). Acta Med Iran. 2016;54(1):39-43. [PubMed ID: 26853289].