1. Background
Meningitis is a life-threatening infection that begins quickly and can be caused by bacterial, viral, or fungal pathogens. Nevertheless, bacterial meningitis is a serious and sometimes fatal disease. Neisseria meningitidis, Streptococcus pneumonia, and Haemophilus influenza type b (Hib) have a leading role (1-5). Since the introduction of the H. influenzae type b conjugate vaccine, detection of N. meningitidis and S. pneumonia has been a priority (6).
In the most recent study on the global burden of disease (GBD), meningitis ranks among the six most burdensome diseases among children under ten (7). In the absence of a comprehensive vaccine for all serotypes and serogroups, laboratories are forced to utilize rapid and high-performance diagnostic methods to combat antimicrobial resistance. The disadvantages of conventional methods, such as low sensitivity and time-consuming, have made these approaches unattractive, while real-time PCR is rapid, sensitive, and multiplicative (8, 9).
2. Objectives
This study aimed to develop a cost-effective biplex real-time PCR to detect N. meningitidis and S. pneumonia through EvaGreen chemistry.
3. Methods
3.1. Bacterial Strains
The bacterial strains of N. meningitidis (ATCC 13090), N. meningitidis (PTCC NO: 1507), S. pneumoniae (ATCC 33400), and S. pneumoniae (ATCC 49619) were used for positive control and analytical sensitivity assessment. Moreover, S. aureus (ATCC 25923), Streptococcus agalactiae (ATCC13813), Escherichia coli (ATCC 25922), Enterococcus faecalis (ATCC 25212), Bacillus cereus (ATCC 9634), H. influenza (ATCC 49766) Listeria monocytogenes, Legionella pneumophila (clinical isolate), Helicobacter pylori (ATCC 26695), Pseudomonas aeruginosa (ATCC 8821), Klebsiella pneumoniae (clinical isolate), Salmonella enterica, Brucella abortus strains S19, 544,133, and RB51, Brucella melitensis strains 16M and Rev-1, Salmonella enterica serovar Typhimurium, Mycobacterium tuberculosis H37Rv (genome isolated previously), and Yersinia enterocolitica were applied to determine the specificity of the assay.
3.2. Nucleic Acid Extraction
DNA extraction was performed by DNAbiotech from a fresh culture of each strain according to the manufacturer’s instructions. Total nucleic acid was eluted in 50μl of elution buffer, and before storage, at -20°C, their quality was monitored by a spectrophotometer (NanoDrop 2000).
3.3. Assay Design
The sequence of target genes for given pathogens was collected from the GeneBank database (NCBI). After alignment, conserved regions of genes of interest were subjected to design primer pairs. AlleleID® software, version 7.75 (PREMIER Biosoft International, Palo Alto, CA, USA), was used for this purpose. Two primer pairs flanked regions of the CtrA (80 bp) and lytA (185 bp) genes specifying N. meningitidis and S. pneumoniae, respectively. Dissociation curve analysis was used to differentiate amplification of N. meningitidis from S. pneumonia.
3.4. EvaGreen Real-Time PCR and Lower Limit of Detection
Samples were run in triplicate on a StepOne Plus (Applied Biosystems) consisting of a heating stage at 95°C for 15 minutes, followed by 40 cycles of denaturation (95°C for 15 seconds), annealing (60°C for 20 seconds), and elongation (72°C for 20 seconds). In order to determine the specificity of each amplification, after each run, a melting curve analysis was conducted. At this stage, the mixture was cooled for 1 min at 60°C and then heated at 95°C for 15 s. Real-time PCR amplifications were performed in a total reaction volume of 25 μL and comprised five μL of HOT FIREPol® EvaGreen® qPCR Mix Plus (Vienna, Austria, Europe), one μL of each primer, and 100 ng of the DNA template. The fluorescent signal is acquired at the extension step in the green channel.
Analytical sensitivity of singleplex and biplex for both pathogens was done by replicate testing of a 7-dilution series of their genomic DNA ranging from 100 - 106 femtogram per reaction. The analytical specificity of the assays was determined by testing related and non-related microorganisms. Separately, the lower limit of detection (LLD) for N. meningitidis and S. pneumonia in the form of singleplex and biplex was determined.
4. Results
4.1. Optimization of the EvaGreen Multiplex PCR
Primer concentrations were optimized by testing 50 to 250 nM, which yielded favorable output at 100. The singleplex assays of every pathogen were compared with their multiplexing in Table 1. Minimal interference was observed among primers of the targets upon multiplexing; hence, we found no cross-reactivity between each primer pair. On the multiplex assay, the reproducibility and repeatability of tested replicates indicated low intra-assay and inter-assay CVs of less than 1.5%.
| EvaGreen Real-Time PCR | Singleplex | Biplex | |
|---|---|---|---|
| Microorganism | Nm | Sp | NM/Sp |
| Ct | 21.93 | 23 | 22/23.11 |
| Tm (°C) | 80.5 | 86.16 | 80.6/86 |
4.2. Analytical Specificity and Sensitivity
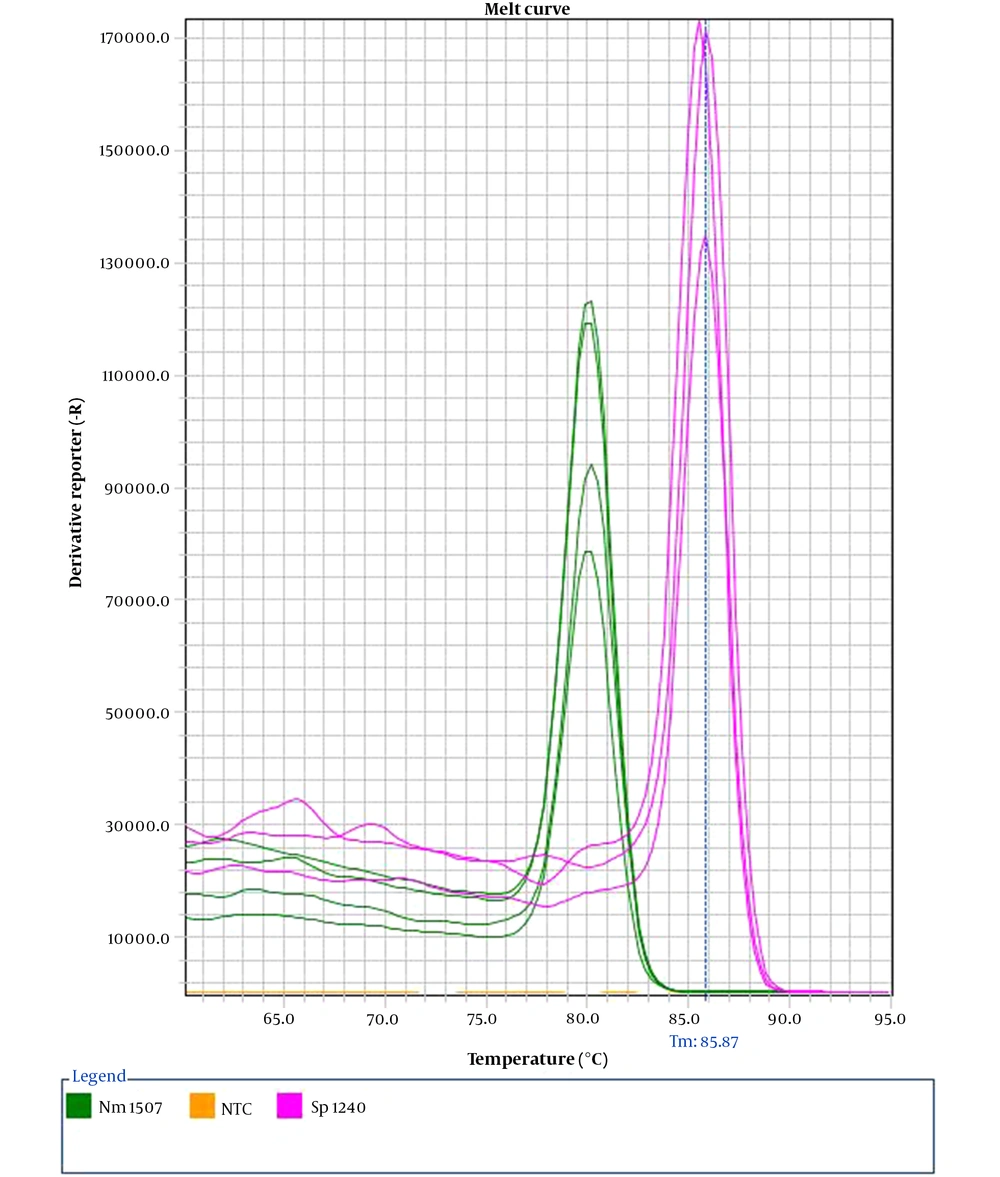
Our assay demonstrated 100% specificity. Exclusive microorganisms did not represent amplification plots; by contrast, inclusive strains were detected in their respective detection channel. Upon multiplexing, we recognize two distinct melt peaks through melt curve analysis, with Tm values of 80.2 ± 0.3°C for N. meningitidis and 85.9 ± 0.2°C for S. pneumoniae (Figure 1).
The LLD of multiplex assay was 20 femtograms equivalent to 9 copies/reaction for N. meningitidis and 30 femtograms equivalent to 13 copies/reaction for S. pneumoniae. These amounts were slightly lower on the singleplex assay than on the biplex, which can be ignored. An analysis of the regression curve of the multiplex assay revealed linearity over a range of concentrations of different microbial genomics with the same R2 value of 0.99 and 100% efficiency for both Nm and Sp. Non-target bacterial reference strains tested negative.
4.3. Diagnostic Validation
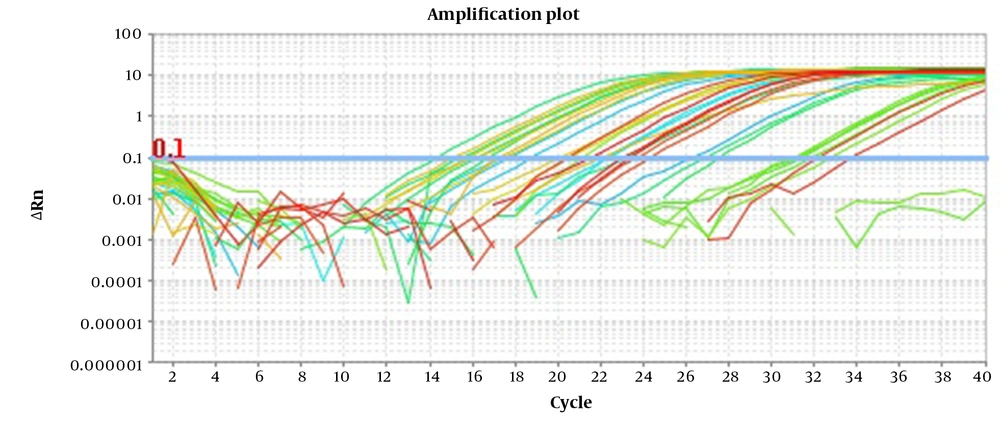
The clinical utility of the assay was evaluated by 51 CSF samples artificially spiked with different amounts of target pathogens. Of these, 23 had Nm, 21 had Sp, and 7 had both Nm and Sp. Moreover, seven negative CSF samples were selected with normal protein and glucose levels and negative culture and gram stain results. After DNA extraction, 5 µl of these samples were tested using multiplex EvaGreen real-time PCR. All three types of positive samples were accurately identified, and no false positive resulted from any negative samples (Figure 2).
5. Discussion
Various bacteria have the potential to cause meningitis; N. meningitidis, S. pneumonia, and H. influenza type b (Hib) have priority. With the advent of H. influenzae type b conjugate vaccines, the importance of pneumococcal and meningococcal meningitis has been exceeded (10-12). Hence, applying rapid, reliable, and cost-effective methods which enable us to identify the abovementioned pathogens in a single tube would be beneficial. As time plays a crucial role in meningitis care, molecular diagnostic tests can pave the way for the right and timely therapy (11).
The importance of culture and gram stain cannot be ignored, as culture is the gold standard. Nevertheless, their insensitivity creates a problem, and it takes time for cultures to be evaluated (13). Moreover, giving antibiotics before hospital admission worsens the situation. Under such circumstances, diagnosis of non-cultured meningitis is of paramount importance to clinical laboratories (14). As a result, a swift and sensitive approach provides a comprehensive epidemiological picture and prevents overtreatment (14).
Although different probe-based multiplex real-time PCR methods are available to detect pathogens, the high cost of dual-labeled fluorescent probe synthesis and additional filter requirements to detect each fluorescent agent present some limitations (15-18). Intercalating dyes such as SYBR Green and EvaGreen, which are affordable, offer an alternative and are widely used. In comparison with SYBR Green, EvaGreen has some advantages. Firstly, it is more sensitive, and secondly, due to its less negative influence on reaction, the occurrence of non-specific amplification is rare (18-20).
In this current study, we designed and developed a multiplex EvaGreen real-time PCR to detect Nm and Sp in a single tube capable of discriminating their amplicon using dissociation curve assessment. The melt curve plot displays a unique peak depending on GC content, length, and sequence. Due to the non-specific nature of EvaGreen, two primer pairs in the same tube may negatively affect each other, whereas our assay produced distinct and specific amplicons and melt curves. Negative controls and other related and unrelated microbes did not exhibit amplification and melt curves.
A sensitive approach is essential for the early detection of pathogens. Our assay can detect 9 and 13 copies/reaction of Nm and Sp, respectively. The study by Kesanopoulos et al. based on SYBR Green revealed a detection limit of 50 copies/reaction for N. meningitidis (14). Comparatively, several studies were also done by SYBR Green and real-Time loop-mediated isothermal amplification technique (LAMP) for detecting Sp and Nm, showing less sensitivity and specificity (21, 22).
Similarly, the strength of agreement between singleplex and duplex in this assay resembles that of studies applying probe-based real-time PCR (8, 23, 24). It is possible, despite different and relatively poor chemistry, that maintaining such strength accounts for the high compatibility and well-designed primer pair. The favorable distance between melt peaks of two amplicons preserves the sensitivity of multiplex assays like singleplex. The formation of primer dimers is a major threat to intercalating dye-based real-time PCR as it may lead to false positive results, especially in multiplex assays (18, 20). Our findings indicated that primer dimer had an insignificant effect on the amplification curve.
5.1. Conclusions
To sum up, EvaGreen-based real-time PCR and melting curve analysis offers a high-throughput and cost-effective method for simultaneous detection of Nm and Sp in CSF samples. Indeed, an efficient diagnosis of meningitis prevents overtreatment and antibiotic resistance.