1. Background
Diarrhea caused by different intestine pathogens is still regarded as one of the problematic complications in public health (1) and bacterial diarrhea has been reported as one of the most common cause of death in developing countries (2). Every year, many patients develop diarrhea around the world including Iran. Among developing countries, bacteria, such as Escherichia, Vibrio, Salmonella and Shigella are the primary microbial agents of diarrhea (3, 4).
Enterotoxigenic Escherichia coli (ETEC), primarily known as the significant cause of non-inflammatory diarrhea, is the cause of diarrhea in 11 to 15% of new born animals and tourists' diarrhea in developing countries. Like other gastrointestinal infection diseases, it is caused by lack of sanitation and most often contamination transfers from contaminated food, water or stool (5, 6). Like other gastrointestinal infectious diseases, poor sanitation as well as contamination transfer from contaminated food, water or stool, are the main causes of diarrheal infection. Heat-labile toxin (LT), heat-stable toxin (ST) and colonization factors (CFs) are suggested as three major virulence factors for ETEC strains, which produce LT, ST or both (7-9). The initial case is the pivotal factor of bacterial virulence, which is vital for ETEC recognition, because it has considerable role in human and animal diarrhea (10). The LT toxin includes A and B subunits. Subunit A is responsible for enzymatic activity and Subunit B is responsible for connecting toxin to the surface of Eukaryotic cells. After the conjunction of LT toxin to the epithelial cells of the small intestine, subunit A is activated through the enhancement of cAMP and finally this mechanism ends with the release of water and electrolytes into the intestine (11). In the past, ETEC reorganization relied on detection of LT or ST enterotoxins. Earlier ST was diagnosed by the means of rabbit ileal loop preparations, yet because of high expenses and ambiguity in standards, infant mice assay has been used instead (12). Also, other experiments like radioimmunoassay (13) and enzyme-linked immunosorbent assay (ELISA) (14) have been done for ST recognition, both of which confirm the results of infant mouse assay. Other diagnostic methods such as radioimmunoassay (13) and enzyme-linked immunosorbent assay (14) have also been utilized for ST recognition, both of which confirmed the results of infant mouse assay. Nowadays, different methods of PCR, especially Multiplex PCR that contain certain primers, are used for recognition of ETEC and its colonization factors (15).
2. Objectives
The aim of this study was detection of LT toxin gene in E. coli clinical strains by PCR-ELISA with specific primers and designed probe.
3. Methods
3.1. Study Design and Sample Society
This experimental and prospective study was conducted on Iranian children communities and stool specimens were obtained from May to November 2014. During the first screening phase, 40 stool samples were gathered from laboratories of different hospitals and care centers of Iran and investigated for defining the presence of ETEC and LT toxin. Inclusion criteria for sampling included (a) loose unformed stools along with (b) vomiting and (c) diarrhea.
Stool samples were gathered on the day of admission before antibiotics therapy. Demographic information such age, gender as well as data related to their symptoms including vomiting, were obtained through questionnaires.
3.2. Bacterial Strains and Microbiological Studies
One ETEC bacteria, which produced LT, was provided by the reference laboratory of Iran and was verified by biochemical and immunologic methods. Furthermore, the two previously isolated ETEC strains from children with diarrhea, characterized for enterotoxin profile, were used as controls of the PCR reaction (16).
Stool specimens were diluted and plated on MacConkey agar (HiMedia, M081B) and then incubated at 37°C for 18 hours. Lactose-fermenting colonies were selected as E. coli and submitted for biochemical tests.
3.3. Genomic DNA Template Preparation by the CTAB Method
Genomic DNA template was prepared by the CTAB method (17). Escherichia coli isolates were cultured in luria-bertani (LB) broth (Sigma, L3522) at 37°C for 16 hours. Freshly prepared culture was centrifuged at 5000 rpm for five minutes. Supernatant fluid was removed and the pellet was suspended in a solution containing 567 µL of TE buffer, 30 µL of 10% SDS, 5 µL proteinase K (Thermofisher, EO0491) and incubated for one hour at 37°C. To this mixture 100 μL of 5M NaCl was added, mixed thoroughly then 80 μL of CTAB/NaCl was added and incubated for 15 minutes at 65°C. An approximately equal volume (0.7 to 0.8 mL) of isoamyl alcohol-chloroform (Merck, 102445) was added, mixed thoroughly and centrifuged at 4°C and 2500 rpm for 25 minutes. The viscous supernatant was transferred to a fresh tube, mixed with an equal volume of chloroform: isoamyl alcohol (24: 1) and centrifuged at 4°C.
DNA from the aqueous phase was extracted with 500 µL of isopropanol and washed with 70% ethanol. The precipitated DNA was resuspended in 100 µL TE buffer containing 50 micrograms of RNase A (Thermofisher, 12091039) and incubated for one hour at 37°C.
3.4. Primer and Probe Design
Considering the fact that the main diarrhea agent is heat-labile toxin (LT), after investigating the bacterial genome, this was the main issue of our first step of diagnosis. For this reason the functional domain (subunit A), which included the enzymatic character, was separated and used for designing a couple of primers for amplification of ltA gene.
The features of the designed primers such as GC content; Tm, ΔG etc. were checked by DNASIS and Oligo software. The primer and probe sequences are presented in Table 1. The oligonucleotides were supplied by SinaClone (IRAN).
| Oligonucleotide | Sequence | Nucleotide Position | Expect product Size (bp) |
|---|---|---|---|
| EsLAF | ATGCCCAGAGGGCATAATGAG | 121 - 141 | 565bp |
| EsLAR | GATATATTGTGCTCAGATTCTGGG | 721 - 776 | |
| EsLAP | GTTTCTGCGTTAGGTGGAA | 391 - 409 |
3.5. Polymerase Chain Reaction
The PCR reaction was performed for amplification of LT gene in a 25 μL total reaction mixture. Each reaction mixture contained (a) 0.4 μm of each primer, (b) 200 μM of dATP, dCTP and dGTP, respectively (c) 190 μM of dTTP, (d) 10 μM DIG-11-dUTP (Roche Diagnostics, 11209256910),(e) 0.5U of Taq DNA polymerase (CinnaGen,TA7506C), (f) 2.5 μL 10X buffer, (g) 2 mM MgCl2 and different concentrations of genomic DNA. The PCR cyclic conditions were initiated at 95°C for five minutes followed by 35 cycles at 95°C for one minute, 63°C for 45 seconds, 72°C for one minute, and a final extension at 72°C for five minutes.
3.6. Detection of Polymerase Chain Reaction products
For gel electrophoresis, a mixture of 5 μL of PCR products and 1 μL of loading buffer was loaded on a well of 1% agarose gel (SinaClone, MR7740C). Furthermore, 100 bp plus DNA ladder was used to determine the size of the PCR products. Electrophoresis was run at 85V for 45 minutes in 0.5X TBE buffer. Finally, the gel was stained with ethidium bromide (Thermofisher, 15585011) and PCR products were visualized under UV light and images were stored by the use of a gel documentation system.
The PCR product was sequenced to confirm the PCR accuracy. For ELISA detection, microtiter wells were coated with one microgram streptavidin (Thermofisher, S866) and kept overnight at 4°C. The wells were washed three times with Phosphate Buffered Saline (PBS) containing 0.05% Tween 20 (Sigma, P1379) (PBST) and blocked using blocking solution containing 3% BSA (Sigma, A9418). After washing, 10 μL of labeled product was added to 90 μL 1X SSC buffer and incubated for ten and five minutes in boiling water and on ice, respectively. In the next step 10 μL of probe was added to the tube. After incubation for two hours at 60°C, 100 μL of this hybridization buffer was added to each well and allowed to remain at 37°C for one hour. This step was followed by a wash with 20% BPST buffer for three times. Anti-digoxigenin antibody conjugated with peroxides (Abcam, ab51949), diluted 1000-fold in Phosphate Buffered Saline with Tween 20 (PBST) buffer, was added to each well in a final volume of 100 µL, and then the plate was incubated at 37°C for one hour. The plates were washed and dried as described earlier. OPD substrate solution (Sigma, P9187) was added to each well and the plate was incubated at room temperature for 10 minutes, in a dark environment. The reaction was stopped with 2.5M H2SO4 and the optical density was measured at 492 nm using an ELISA reader (Dynex Technologies, Guornesey, Channel Islands and Great Britain). The values represent the mean of three experiments in duplicate.
3.7. Sensitivity and Specificity of Polymerase Chain Reaction-Enzyme Linked Immunosorbent Assay
To determine the detection limit for LT toxin gene, genomic DNA was extracted and serial dilutions of the extracted DNA was used for PCR-ELISA. The specificity of the PCR-ELISA was determined using genomic DNA of collected samples from children and non Enterotoxigenic E. coli, Klebsiella pneumonia, Pseudomonas aeruginosa, Salmonella typhimurium, Vibrio cholera, and Shigella dysenteriae. The PCR was carried out according to the mentioned protocols and the products were analyzed by agarose gel electrophoresis and ELISA.
3.8. Detection of Clinical Samples by Polymerase Chain Reaction-Enzyme Linked Immunosorbent Assay
In this study, 40 positive samples of E. coli obtained from stool were analyzed. Stool sample cultures were collected from the laboratory of different hospitals and day care centers before initiation of any antimicrobial therapy. The samples were cultured in LB medium and chromosomal DNA was prepared. After determining the optical density (OD) at 260 nm, PCR was performed according to the previous plan in 35 cycles. PCR-ELISA was carried out.
The values represent the mean of three experiments in duplicate.
3.9. Statistical Analysis
Data were reported as mean ± SD and statistical analysis was performed using one-way analysis of variance (ANOVA) (SPSS 16.0). The significance (P < 0.010) of differences was assessed by post hoc comparison of means using lowest significant differences (Dunkan).
4. Results
4.1. Polymerase Chain Reaction detection of LT A gene
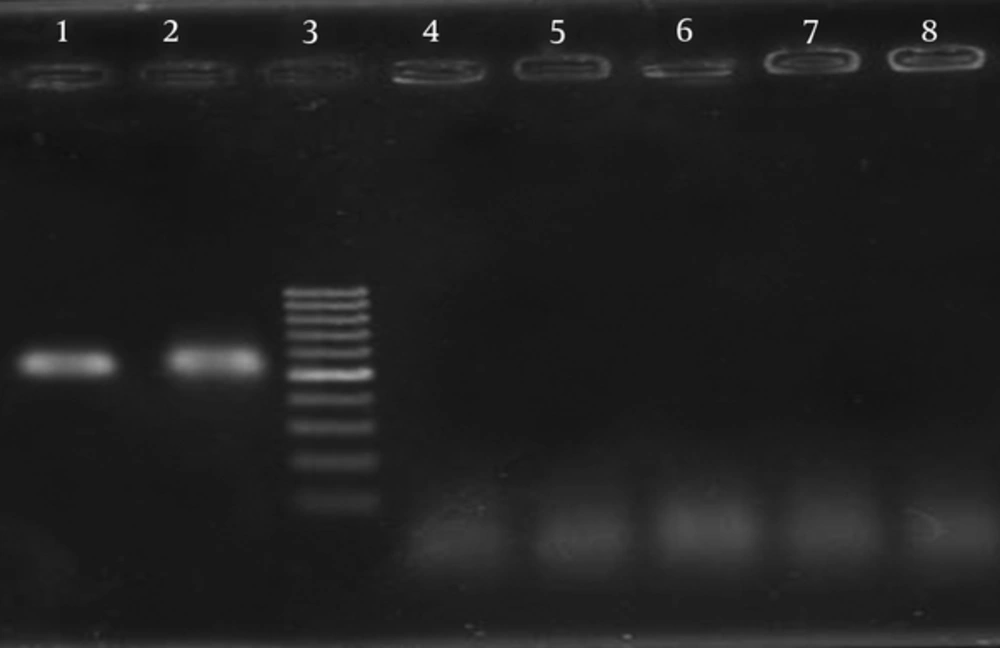
The PCR primers and a capture probe were designed based on the LTA gene encoding A subunit of LT toxin from ETEC. The specificity of the primers was tested with DNA extracted from ETEC as positive control and non-ETEC strains as negative controls. Agarose gel electrophoresis of extracted DNA showed that genomic DNA in large quantities and of good quality was obtained. The purity of the DNA samples was confirmed by absorbance (A260/A280) ratio, which was 1.8 to 2.0. Several conditions such as various annealing temperatures (57 to 63°C), different concentrations of MgCl2, dNTPs and primers were examined for optimization of PCR reaction. In our examination, 63°C was recognized as the best temperature, which led to amplification of 565 bp PCR products of LT A gene (Figure 1).
Sequencing of PCR product showed that PCR program amplified specific target using the designed primer.
For the labeling of digoxigenin, a PCR reaction was performed with digoxigenin labeling mix and the results were analyzed on the 1% agarose gel (Figure 1).
4.2. Specificity and Sensitivity of Polymerase Chain Reaction
The standard PCR with specific primers was done for ETEC, Salmonella typhimurium, Klebsiella pneumonia, Pseudomonas aeruginosa, Vibrio cholera and Shigella dysenteriae.
The PCR products of the expected size of 565 bp were amplified for ETEC strain, and no products were amplified for any of the negative control strains (Figure 1). Serial dilution of extracted DNA was prepared and PCR reactions were performed on each diluted DNA sample. The amplification products were examined by gel electrophoresis. Regarding the concentration of primary sample, sensitivity of the reaction was calculated as 190 pg/μL.
4.3. Polymerase Chain Reaction-Enzyme Linked Immunosorbent Assay Specificity and Sensitivity Assay
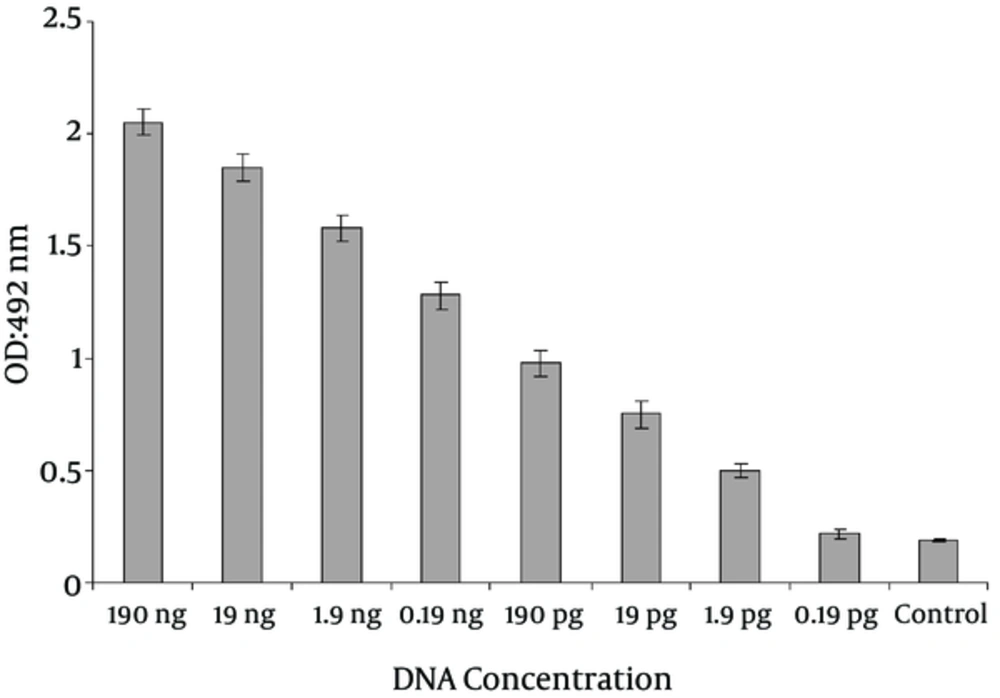
For determining the sensitivity of PCR-ELISA, serial dilutions of genomic DNA were prepared and for confirming the PCR products of LT gene, the hybridization method was used. The detection limit of PCR-ELISA on ETEC was determined to be as low as 1.9 pg (Figure 2). A statistically significant difference in detection limit of LT toxin gene between PCR and PCR- ELISA was observed (P < 0.010).
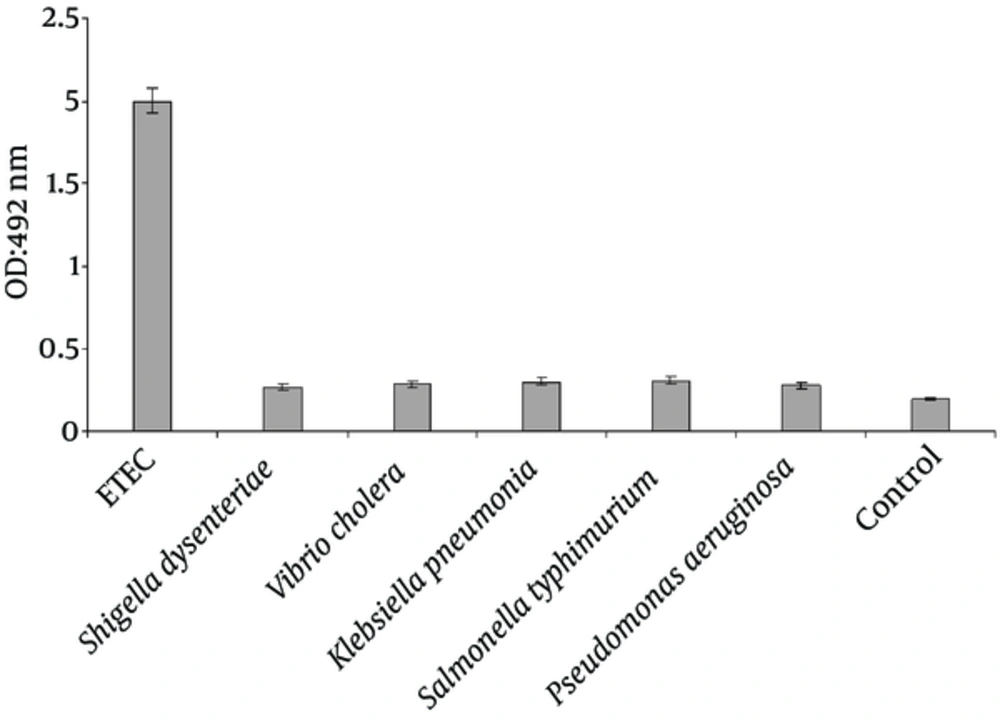
The specificity of the PCR-ELISA was analyzed using genomes of Shigella dysenteriae, Vibrio cholera, Klebsiella pneumonia, Salmonella typhimurium and Pseudomonas aeruginosa (Figure 3).
Optical density of PCR-ELISA reaction from ETEC strain significantly increased compared with other bacterial strains (P < 0.001). Moreover, there was no significant difference between Optical density of PCR-ELISA reaction for Shigella dysenteriae, Vibrio cholera, Klebsiella pneumonia, Salmonella typhimurium and Pseudomonas aeruginosa strains (P < 0.001).
4.4. Clinical Samples Analysis
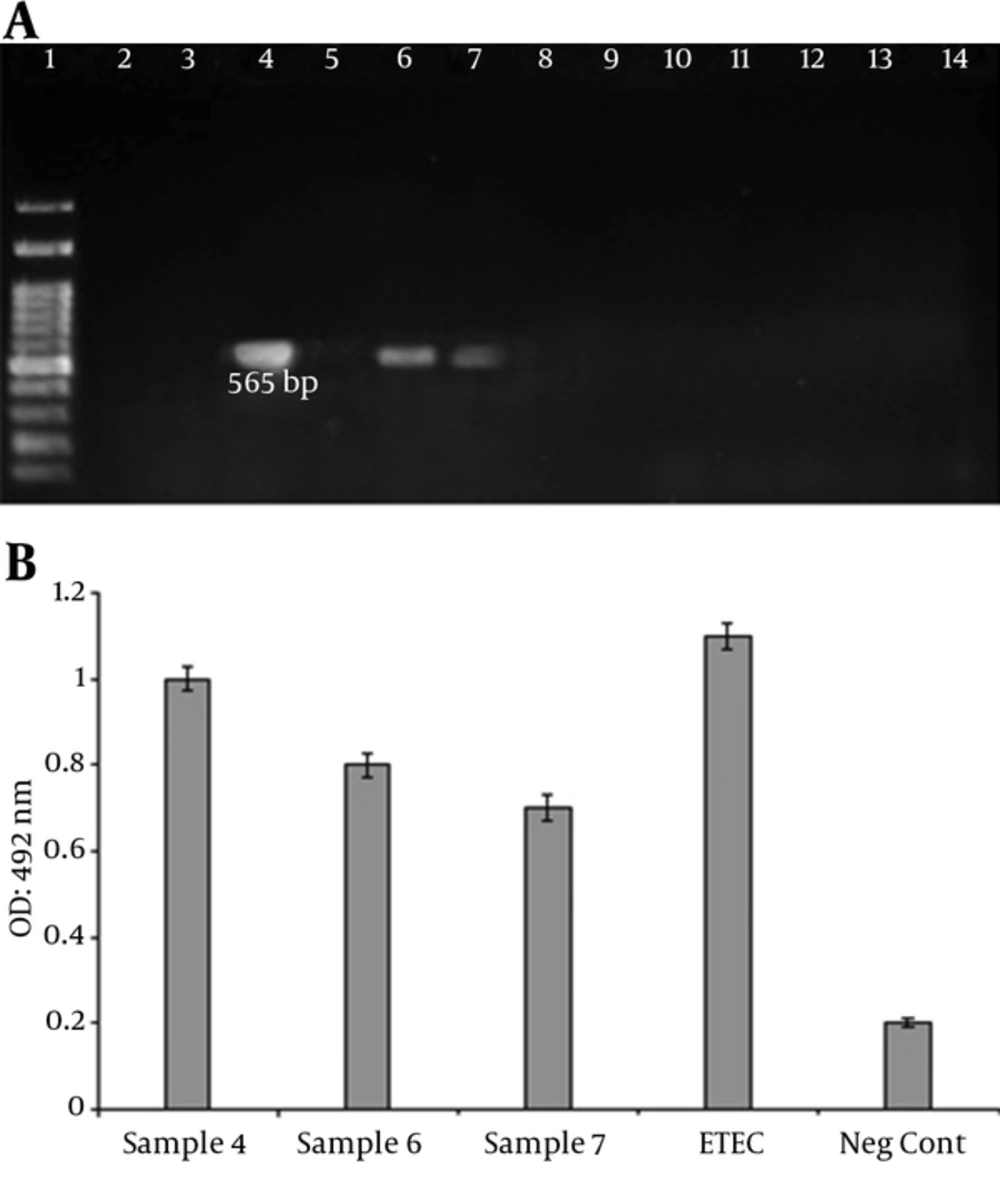
A total of 40 clinical samples were collected and screened for the presence of ETEC strains. Three (7.5%) of the samples were detected as LT positive (Figure 4). Age distribution among patients was from < 1 to 40 years of age. The ETEC affected patients with an average age of eight years, with 75% (2 Nos.) less than ten years of age. The most common symptoms were Watery stool and abdominal pain. Bloody diarrhea was not observed in the patients. Gender distribution of the patients with ETEC was two females (75%) and one male (25%).
5. Discussion
Nowadays diarrheal disease is the most frequent illness around the world. Diarrhea can be caused by many factors including bacterial and viral agents.
Furthermore, ETEC is one of the most common bacteria responsible for diarrhea in different parts of the world (18). The ETEC bacteria threaten the lives of many people, especially children under five years old all around the world and also in Iran (4, 5, 19). Detection of ETEC relies on the identification of enterotoxins. Several Genotypic and phenotypic methods were used for detecting LT toxins in various samples (20). Rabbit ileal loop model was the golden standard method for identification of LT toxin from ETEC strains. The Rabbit ileal loop assay is time consuming and requires skilled personnel and it also yields variable results. The ELISA technique has been used for detection of LT toxin using GM1 ganglioside as a receptor, and is based on monoclonal antibodies against LT (21). Need for sample cultures, lower sensitivity and time consuming issues are among the disadvantages of GM1-ELISA compared with polymerase chain reaction (22). In epidemiological studies, toxin expression might have been lost during culture of clinical and GM1-ELISA could not detect these strains that initially expressed LT toxin in patients (20). Agglutination tests are available for detecting LT enterotoxin. This method is simple, easy and rapid but is based on monoclonal antibodies and therefore requires expensive facilities (23). Serotyping method has also been used to identify and characterize ETEC strains but since more than 112 O and H groups have been identified among ETEC, this method is difficult (24). However, sensitive and specific molecular techniques have been developed and are in many cases preferable due to reduced time of the tests implementation (25).
Genotypic methods targeting virulence genes such as PCR are becoming the standard procedure. DNA probes can be used for the detection of LT-encoding genes in various samples such as stool and environmental samples. Several sensitive and specific PCR assays exist for ETEC detection (16).
Real time PCR and microarrays with the possibility to quantify the bacteria in the sample has been introduced in ETEC research. However, these methods are expensive and laboratories in developing countries cannot afford routine analysis (26).
Blanco et al. and Rajkhowa et al. (27, 28) by the means of PCR and specialized primers, which were designed for LT gene diagnosed LT toxin of ETEC in calves. Shahrani et al. (29) used molecular PCR method for the detection of ETEC. Their study was based on diagnosing ST and LT genes. Nazarian et al. (16) used Multiplex PCR reaction for detection ETEC toxin and colonization factors.
In our research, we used PCR-ELISA for fast detection of ETEC. The PCR-ELISA method detects nucleic acid instead of protein and is much more sensitive compared to conventional PCR assays, with lower detection limit and shorter analytical time (30). The most basic step in designing a diagnostic reaction is the selection of suitable fragments and designing necessary primers for the purpose of segments duplication. Other researchers used PCR-ELISA for recognizing representative coliforms in water samples (31), and direct identification of Pseudomonas aeruginosa from positive BACTEC blood culture bottles. Mousavi et al. (32) evaluated a PCR-enzyme-linked immunosorbent assay for sensitive and rapid detection of V. cholera O1 from Iran. In our previous study, PCR-ELISA was used to detect genes encoding shiga toxins1 and 2 from Escherichia coli O157: H7 and other Shiga toxin-producing E. coli (STEC) (33). In our procedure, we used DNA genomic ETEC; we also decreased the number of PCR cycles to five for reducing the reaction time. Using specifically designed primers and a capture probe, the PCR-ELISA method described above was sensitive enough to detect 1.9 pg/µL of ETEC genome, and this level of detection was achieved within three hours. Specificity of this method with other strains of bacteria was examined and the results showed that all the primers and probes, which were used in this research, were only assigned to ETEC. Our data indicated that PCR-ELISA is highly specific, and its usage, instead of conventional gel electrophoresis, could be able to increase the sensitivity of the assay. By offering faster diagnostic time and high sensitivity, there is high potential for PCR-ELISA to serve as a powerful detection tool in medical, food and agriculture fields. The results of the study show that PCR–ELISA method is sensitive, specific and rapid for detection of ETEC.