Abstract
Background:
In the past two decades, the incidence of fungal infections has significantly increased worldwide. Despite treatment with a broad range of antifungal agents, nosocomial candidemia is associated with high mortality rates and resistance to antifungal agents is becoming increasingly prevalent in Candida species. Therefore, detection and identification of pathogens at the species level and antifungal susceptibility testing are essential to select the appropriate antifungal therapy. The incidence of candidemia and the antifungal susceptibility patterns of the associated strains have not been extensively studied in Iran.Objectives:
The current study aimed to assess the microbial epidemiology of candidemia and the antifungal susceptibility profiles of Candida isolates.Patients and Methods:
Out of 5141 blood culture specimens analyzed in Iran, 48 specimens from 32 patients were yeast-positive. The isolates were precisely identified at the species level using the well-established phenotypic polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) method. The yeasts were also tested for antifungal susceptibility to fluconazole, itraconazole, ketoconazole, voriconazole, and caspofungin using the broth microdilution method.Results:
Candidaparapsilosis (C. parapsilosis) was the most common yeast pathogen isolated (34.4%), followed by Candidaglabrata (C. glabrata) (28.1%), Candidaalbicans (C. albicans) (25%), Candidatropicalis (C. tropicalis) (9.4%), and Candidakefyr (C. kefyr) (3.1%). Candidaglabrata was isolated more often in elderly patients (> 60 years old).Conclusions:
The results of antifungal susceptibility tests demonstrated that voriconazole was the most active drug against Candida isolates. Candida albicans is the most common yeast isolated from human blood world-wide; the unexpectedly higher rate of C. parapsilosis isolated in this study necessitates more studies with larger sample sizes in Iran.Keywords
Polymerase Chain Reaction Polymorphism Restriction Fragment Length Candida parapsilosis Voriconazole Fungemia
1. Background
In the past two decades, the incidence of fungal infections has significantly increased worldwide due to the increased use of chemotherapeutic and immunosuppressive agents, and organ transplantation (1). Despite treatment with a broad range of antifungal agents, nosocomial candidemia is associated with high mortality rates (60% - 80%), especially in patients in intensive care units (ICUs), and immunocompromised patients (2). More than 100 yeast species have been identified as pathogens, but fewer than ten are common causes of fungal infections in humans. In general, Candida spp. are the most prevalent fungal pathogens, and C. albicans is the most commonly isolated species from patients with candidemia. Nevertheless, other Candida species have emerged as clinically important opportunistic pathogens (3-6). Resistance to antifungal agents is becoming increasingly prevalent in Candida species (7). For example, C. lusitaniae, C. guilliermondii, and C. kefyr have been reported as resistant to amphotericin B (8). Resistance can increase the risk of treatment failure, mortality and prolonged hospital stays (9). Therefore, detection and identification of pathogens at the species level and antifungal susceptibility testing are essential to select the appropriate antifungal therapy, particularly for patients with invasive fungal infections. The incidence of candidemia and the antifungal susceptibility patterns of the associated strains have not been extensively studied in Iran. The current study aimed to evaluate the prevalence of Candida spp. and their susceptibility profiles in blood isolates of Iranian patients. Polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP), as well as more conventional methods, was used to identify Candida species isolated from blood culture specimens. In addition, the susceptibility profiles of clinical isolates against current antifungal agents were determined.
2. Objectives
The current study aimed to assess the microbial epidemiology of candidemia and the antifungal susceptibility profiles of Candida isolates among hospitalized patients.
3. Patients and Methods
3.1. Clinical Specimens
A total of 5141 blood samples from patients in Iran were cultured during 15 months. The yeasts isolated from the cultures with significant colony counts were sub-cultured on Sabouraud agar and incubated at 37°C for 2-3 days. Colonies were then transferred into tubes containing 30% glycerol in distilled water and kept at -20°C for further study.
3.2. Morphological Identification of Yeast Isolates
The yeasts were first identified according to morphological characteristics and physiological properties using techniques such as germ tube test, the ability to produce chlamydoconidia, color formation in chromagar Candida, and API ID 32°C (Biomeriuex, Marcy l'Etoile, France).
3.3. Identification of Candida Species by PCR-RFLP
After preliminary morphological identification, yeasts DNAs were isolated using FTA® Elute MicroCards (Whatman Inc., Clifton, NJ, USA) in accordance with the manufacturer’s instructions with minor modifications. Briefly, a loopful of fresh colonies were suspended in 100 µL of sterile water, 4 µL of each suspension was dropped on a 3 mm diameter FTA card disc, and the discs were incubated at room temperature for at least three hours. The dried paper discs were eluted in 500 µL distilled water for a few seconds, transferred to a new tube containing 30 µL distilled water, and incubated at 94°C for 20 minutes. They were then removed and the water containing the DNA was used for PCR or stored at -20°C until required. The Candida isolates were identified according to a method described by Mirhendi et al. (10). Briefly, universal primers ITS1 (5'-TCCGTAGGTGAACCTGCGG- 3’) and ITS4 (5’-TCC TCC GCT TAT TGA TAT GC-3´) were used to allow amplification of the ITS1-5.8 SrRNA-ITS2 regions. PCR amplification was carried out in a final volume of 50 µL; each reaction contained 1 µL template DNA, 0.5 µM of the primers, 0.40 mM of deoxynucleoside triphosphate (dNTP), 5 µL of 10x PCR buffer and 1.25 U of Taq DNA polymerase. An initial denaturation step at 94°C for 5 minutes was followed by 35 cycles of denaturation at 94°C for 30 seconds, annealing at 56°C for 45 seconds with an extension at 72°C for one minute, and a final extension step at 72°C for seven minutes. The presence of specific 400-900 base-pair PCR products was examined by staining with ethidium bromide after electrophoresis on 1.5% agarose gel. The amplified PCR products were digested with the restriction enzyme MspI (Thermo Fisher Scientific, Waltham, Massachusetts, USA) to differentiate common yeast species including C. albicans, C. tropicalis, C. glabrata, C. parapsilosis, Candida krusei (C. krusei) and C. guilliermondii (10). As there is no cutting site in the internal transcribed spacer (ITS) region of C. parapsilosis, the enzyme ClaI (Roche Molecular) was used to reconfirm these isolates (Figure 1).
The Diagram of PCR-RFLP Routes for Identification of Clinical Candida Isolates in This Study
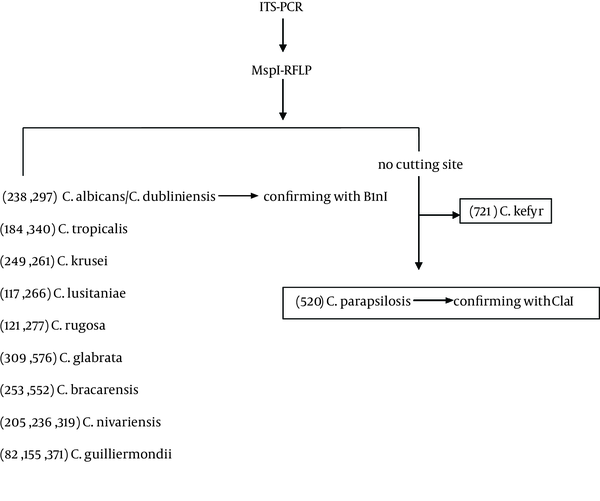
In addition, B1nI (AvrII) (Roche Molecular, Basel, Switzerland) was used to differentiate Candida dubliniensis (C. dubliniensis) from C. albicans. To digest PCR products, 10 µL of the product was incubated with 10 U of each enzyme in a final reaction volume of 25 µL at 37°C for 2 - 3 hours. The digests were examined by staining with ethidium bromide and 2% agarose gel electrophoresis and the yeast was identified according to the restriction fragment patterns for each species.
3.4. Antifungal Susceptibility Testing
Antifungal susceptibility to fluconazole, itraconazole, ketoconazole, voriconazole and caspofungin was tested using the broth microdilution method as recommended by the Clinical and Laboratory Standards Institute (CLSI, document M27-A2). Selected antifungal dilutions were prepared in 3-(N-morpholino) propanesulfonic acid (MOPS)-buffered RPMI-1640 in 96-well microtiter plates. The tested concentration ranges were from 0.125 to 256 μg/mL for fluconazole, and from 0.016 to 32 μg/mL for the other drugs (11). After overnight growth on sabouraud dextrose agar at 35°C, each Candida isolate was suspended in 5 mL of sterile water and vigorously vortexed. The turbidity was adjusted to 0.5 McFarland standard (approximately 1 × 106 to 5 × 106 CFU/mL). The inoculum was diluted to 0.5 × 103 - 0.25 × 103 CFU/mL with RPMI-1640 medium. The microtiter plates were inoculated with 100 µL aliquots and incubated at 35°C for 24 - 48 hours in a humid atmosphere. Growth controls containing drug-free RPMI-1640 medium were included for each isolate. In addition, 200 μL of uninoculated RPMI-1640 medium was included as a sterile control and blank. The MICs were visually determined and defined as the lowest drug concentration at which a prominent decrease in turbidity was observed, compared with that of the drug-free growth-control well. The MICs for amphotericin B were determined as the lowest concentration at which no visible growth was detected. Each experiment was run in triplicate. The American Type Culture Collection (ATCC) type strains including C. albicans ATCC 10261, C. tropicalis ATCC 750 and C. krusei ATCC 6258 were also tested as quality controls.
4. Results
In total, 48 out of 5141 (0.93%) blood cultures were positive for yeasts. These blood samples had been derived from 32 patients (20 males and 12 females, aged 25 - 50 years old). Five patients had two positive blood cultures each, three patients had three positive cultures each, and two patients had four positive cultures each. Predisposing conditions included cancer (n = 6), surgery (n = 6), haematological malignancy (n = 4), diabetes (n = 2), renal failure (n = 3), pancreatitis (n = 1) and premature birth (n = 1) (Table 1).
| Underlying Disease, No. | Gender | Age | Candida species, No. | Mortality Rate | |
|---|---|---|---|---|---|
| Male | Female | ||||
| Cancer, 6 | 4 | 2 | 59 ± 14 | C. glabrata, 5; C. parapsilosis, 1 | 2 |
| Diabetes, 2 | 1 | 1 | 57 ± 16 | C. albicans, 1; C. parapsilosis, 1 | |
| Surgery, 6 | 4 | 2 | 59 ± 19 | C. albicans, 4; C. parapsilosis, 2 | 1 |
| Haematological malignancy, 4 | 2 | 2 | 14 ± 13 | C. parapsilosis, 4 | |
| Pancreatitis, 1 | 0 | 1 | 47 | C. albicans, 1 | |
| Premature birth, 1 | 1 | 0 | C. albicans, 1 | ||
| Renal failure, 3 | 1 | 2 | 62 ± 15 | C. albicans, 1; C. glabrata, 1; C. parapsilosis, 1 | 1 |
| Unknown, 8 | 6 | 1 | 75 ± 5 | C. albicans, 2; C. glabrata b, 3; C. parapsilosis, 2; C. tropicalis b, 2 | 1 |
| 1 | 0 | 3.5 | C. kefyr, 1 | ||
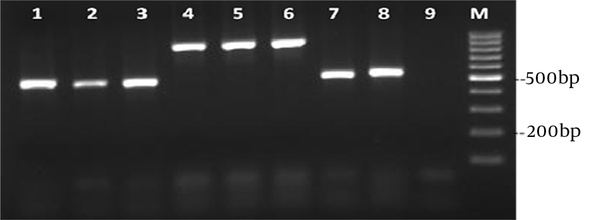
Six patients did not survive; two of them were infected with C. albicans, two with C. parapsilosis and two with C. glabrata (Table 1). The PCR amplification of DNA was successful for all Candida isolates, providing PCR products of approximately 400-900 nucleotides in length (Figure 2).
Agarose gel Electrophoresis of PCR Products to Identify Yeast Blood Isolates
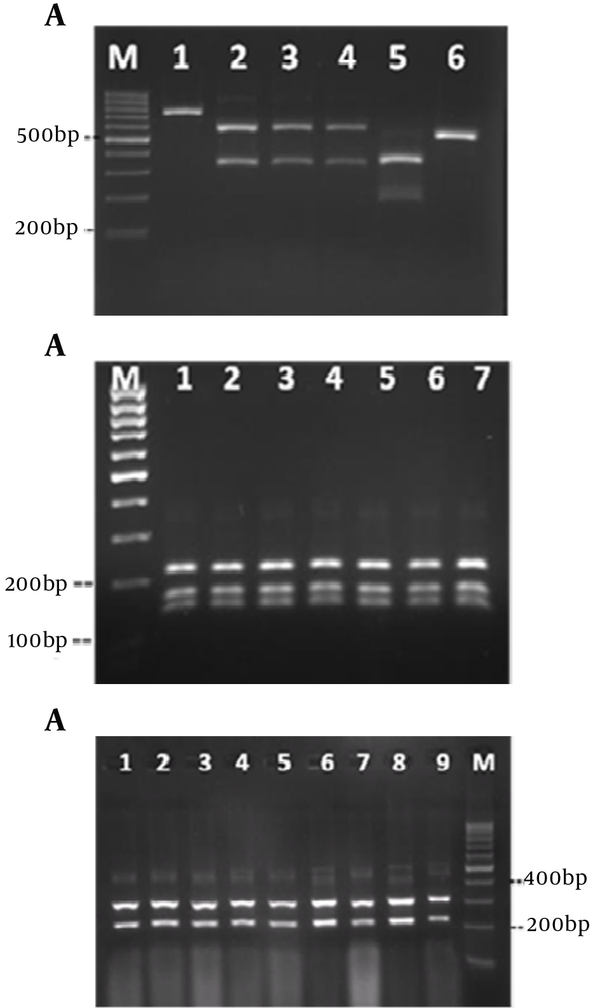
DNA cleavage was performed by the restriction enzyme MspI (Figure 3A). The sizes and patterns of the fragments obtained from the clinical isolates were compared with the patterns from the reference strains. Digestion of the ITS region of Candida species by MspI generated two bands for C. albicans, C. glabrata and C. tropicalis. However, there were no recognition sites on the ITS region of C. parapsilosis. Thus, the RFLP product of C. parapsilosis was the same as the corresponding PCR product (Figure 3A). Identification of C. albicans and C. parapsilosis were confirmed by the restriction enzymes MboI and ClaI, respectively (Figures 3 B and 3 C).
Enzyme Digestion of Amplified Candida DNA
Application of a combination of conventional methodology and PCR-RFLP indicated that the most commonly isolated species was C. parapsilosis (n = 11, 34.4%), followed by C. glabrata (n = 9, 28.1%) and C. albicans (n = 8, 25%). C. tropicalis (n = 3, 9.4%) and C. kefyr (n = 1, 3.1%) were only occasionally isolated (Table 2).
| Isolates, n = 48, No. (%) | Patients, n = 32, No. (%) | |
|---|---|---|
| C. parapsilosis | 17 (35.4) | 11 (34.4) |
| C. glabrata | 18 (37.5) | 9 (28.1) |
| C. albicans | 9 (18.8) | 8 (25) |
| C. tropicalis | 3 (6.3) | 3 (9.4) |
| C. kefyr | 1 (2.1) | 1 (3.1) |
One patient yielded both C. glabrata and C. tropicalis in the blood culture, indicating a double infection. All patients with haematological malignancy were infected with C. parapsilosis, while five patients with cancer were infected with C. glabrata (Table 1). Candida glabrata, the second most commonly isolated Candida species, was found more frequently in elderly patients (> 60 years old) than in the young candidates (Figure 4).
Incidence of Candida Species Isolated From Blood of Patients With Candidemia by Age (Year)
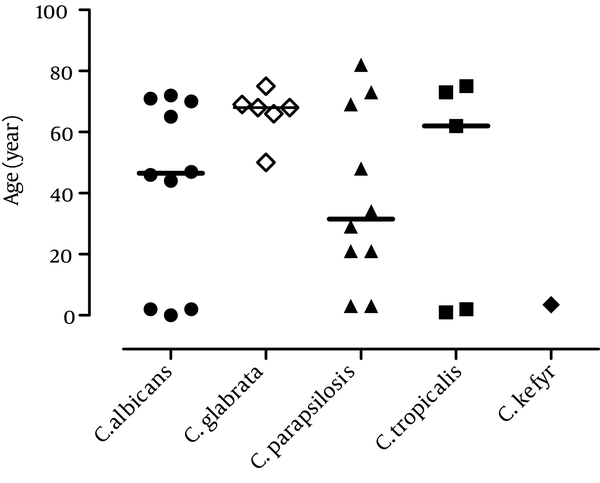
4.1. Antifungal Susceptibility of Clinical Candida Species
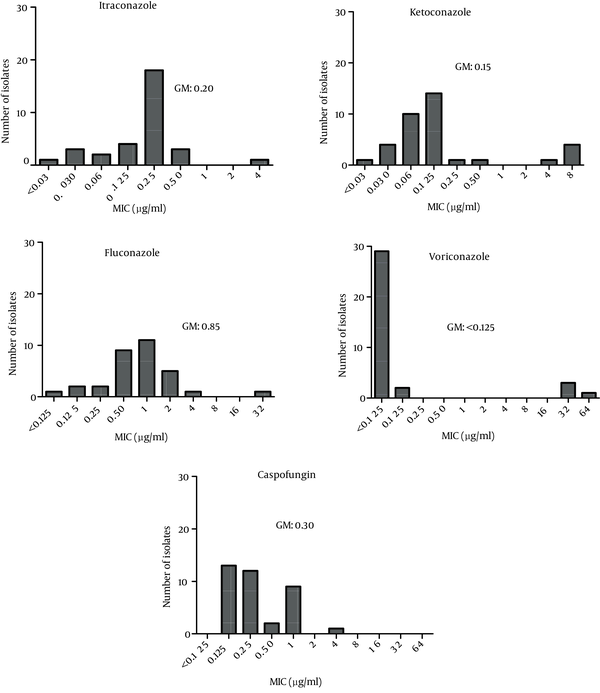
The MIC ranges and geometric means (GMs) obtained in the antifungal tests are summarized in Figure 5. Voriconazole was the most active agent against isolated Candida species (GM < 0.125). The highest GM for the MIC results was obtained for fluconazole (GM = 0.85). Caspofungin was associated with a wide range of MICs (0.125 - 4), with a GM of 0.30.
MICs of Itraconazole, Ketoconazole, Fluconazole, Voriconazole and Caspofungin to Clinical Specimens of Candida spp. Obtained From Blood Specimen of Patients With Candidemia
5. Discussion
In the current study, the yeast strains isolated from blood culture specimens of the patients with candidemia were identified at the species level by conventional and molecular (PCR-RFLP) methods. This was the first study investigating the prevalence of Candida species in positive blood cultures of Iranian patients. The recent increase in infections caused by non-albicans Candida species indicates the need for freely available and rapid tests to identify these species (12, 13). PCR-RFLP was able to reliably identify C. parapsilosis, C. glabrata, C. albicans and C tropicalis, which were the most clinically relevant Candida species. Surprisingly, the current study found that most cases of candidemia were caused by C. parapsilosis (34.4%) and C. glabrata (28.1%). Historically, C. albicans and C. glabrata were reported to be the most frequently identified yeasts isolated from blood cultures worldwide; however, in the recent years, the proportion of infections caused by species other than C. albicans has markedly increased and C. parapsilosis is now one of the leading causes of invasive candidal disease (5, 14, 15). The current study’s findings are in agreement with those of Das et al. who recently reported that 58% of episodes of candidemia were caused by non-albicans Candida (16). In accordance with the current study, Arslankoylu et al. found that C. parapsilosis was the most common Candida species in patients with candidiasis in a paediatric intensive care unit in 2011 (17).
The current study found that C. glabrata was only isolated from middle-aged or older patients, which was in agreement with the findings of Malani et al. who found that C. glabrata fungaemia was most often observed in older adults (18). The relationship between isolation of Candida species and the underlying condition was also studied. Surgery, cancer, haematological malignancy, diabetes, renal failure, premature birth and pancreatitis were the predisposing risk factors for candidemia in the current study. Six patients (of 32, 18.8%) with candidemia did not survive the infection; this is a lower figure than what was reported by Das et al. who found a crude mortality rate of 32% in patients with candidemia in the United Kingdom. This could reflect the fact that C. albicans was the most common Candida species isolated in their study, but could also be the result of differences in age and the underlying conditions (16). Antifungal susceptibility tests demonstrated that voriconazole was the most active drug against isolated Candida species, which is in agreement with a previous study by Labbe et al. who used the CLSI broth microdilution method (19). The current study’s results determined that fluconazole had the highest MIC GM, which is also in agreement with that of Labbe et al., who found the highest MIC for fluconazole compared with various other azole antifungal drugs (fluconazole, voriconazole, ravuconazole, itraconazole and posaconazole) (19). In conclusion, although C. albicans is the most common blood isolate of yeasts world-wide, C. parapsilosis was the most commonly isolated yeast in the current study; this was an unexpected finding that necessitates more studies with larger sample sizes in Iran.
Acknowledgements
References
-
1.
Sipsas NV, Lewis RE, Tarrand J, Hachem R, Rolston KV, Raad II, et al. Candidemia in patients with hematologic malignancies in the era of new antifungal agents (2001-2007): stable incidence but changing epidemiology of a still frequently lethal infection. Cancer. 2009;115(20):4745-52. [PubMed ID: 19634156]. https://doi.org/10.1002/cncr.24507.
-
2.
Gudlaugsson O, Gillespie S, Lee K, Vande Berg J, Hu J, Messer S, et al. Attributable mortality of nosocomial candidemia, revisited. Clin Infect Dis. 2003;37(9):1172-7. [PubMed ID: 14557960]. https://doi.org/10.1086/378745.
-
3.
Chen LY, Liao SY, Kuo SC, Chen SJ, Chen YY, Wang FD, et al. Changes in the incidence of candidaemia during 2000-2008 in a tertiary medical centre in northern Taiwan. J Hosp Infect. 2011;78(1):50-3. [PubMed ID: 21316800]. https://doi.org/10.1016/j.jhin.2010.12.007.
-
4.
Rangel-Frausto MS, Wiblin T, Blumberg HM, Saiman L, Patterson J, Rinaldi M, et al. National epidemiology of mycoses survey (NEMIS): variations in rates of bloodstream infections due to Candida species in seven surgical intensive care units and six neonatal intensive care units. Clin Infect Dis. 1999;29(2):253-8. [PubMed ID: 10476721]. https://doi.org/10.1086/520194.
-
5.
Zirkel J, Klinker H, Kuhn A, Abele-Horn M, Tappe D, Turnwald D, et al. Epidemiology of Candida blood stream infections in patients with hematological malignancies or solid tumors. Med Mycol. 2012;50(1):50-5. [PubMed ID: 21696259]. https://doi.org/10.3109/13693786.2011.587211.
-
6.
Cohen Y, Karoubi P, Adrie C, Gauzit R, Marsepoil T, Zarka D, et al. Early prediction of Candida glabrata fungemia in nonneutropenic critically ill patients. Crit Care Med. 2010;38(3):826-30. [PubMed ID: 20042858]. https://doi.org/10.1097/CCM.0b013e3181cc4734.
-
7.
Metin DY, Hilmioglu-Polat S, Samlioglu P, Doganay-Oflazoglu B, Inci R, Tumbay E. Evaluation of antifungal susceptibility testing with microdilution and Etest methods of Candida blood isolates. Mycopathologia. 2011;172(3):187-99. [PubMed ID: 21424603]. https://doi.org/10.1007/s11046-011-9413-y.
-
8.
Amran F, Aziz MN, Ibrahim HM, Atiqah NH, Parameswari S, Hafiza MR, et al. In vitro antifungal susceptibilities of Candida isolates from patients with invasive candidiasis in Kuala Lumpur Hospital, Malaysia. J Med Microbiol. 2011;60(Pt 9):1312-6. [PubMed ID: 21459913]. https://doi.org/10.1099/jmm.0.027631-0.
-
9.
Lewis RE. Overview of the changing epidemiology of candidemia. Curr Med Res Opin. 2009;25(7):1732-40. [PubMed ID: 19519284]. https://doi.org/10.1185/03007990902990817.
-
10.
Mirhendi H, Bruun B, Schonheyder HC, Christensen JJ, Fuursted K, Gahrn-Hansen B, et al. Differentiation of Candida glabrata, C. nivariensis and C. bracarensis based on fragment length polymorphism of ITS1 and ITS2 and restriction fragment length polymorphism of ITS and D1/D2 regions in rDNA. Eur J Clin Microbiol Infect Dis. 2011;30(11):1409-16. [PubMed ID: 21607825]. https://doi.org/10.1007/s10096-011-1235-9.
-
11.
Rex JH, Alexsander BD, Andes D, Arthington-Skaggs B, Brown SD, Chaturvedi V, et al. Reference method for broth dilution antifungal susceptibility testing of yeasts. Clinical and Laboratory Standards Institute. 2008;28(14).
-
12.
Hachem R, Hanna H, Kontoyiannis D, Jiang Y, Raad I. The changing epidemiology of invasive candidiasis: Candida glabrata and Candida krusei as the leading causes of candidemia in hematologic malignancy. Cancer. 2008;112(11):2493-9. [PubMed ID: 18412153]. https://doi.org/10.1002/cncr.23466.
-
13.
Horn DL, Neofytos D, Anaissie EJ, Fishman JA, Steinbach WJ, Olyaei AJ, et al. Epidemiology and outcomes of candidemia in 2019 patients: data from the prospective antifungal therapy alliance registry. Clin Infect Dis. 2009;48(12):1695-703. [PubMed ID: 19441981]. https://doi.org/10.1086/599039.
-
14.
Playford EG, Marriott D, Nguyen Q, Chen S, Ellis D, Slavin M, et al. Candidemia in nonneutropenic critically ill patients: risk factors for non-albicans Candida spp. Crit Care Med. 2008;36(7):2034-9. [PubMed ID: 18552700]. https://doi.org/10.1097/CCM.0b013e3181760f42.
-
15.
Trofa D, Gacser A, Nosanchuk JD. Candida parapsilosis, an emerging fungal pathogen. Clin Microbiol Rev. 2008;21(4):606-25. [PubMed ID: 18854483]. https://doi.org/10.1128/CMR.00013-08.
-
16.
Das I, Nightingale P, Patel M, Jumaa P. Epidemiology, clinical characteristics, and outcome of candidemia: experience in a tertiary referral center in the UK. Int J Infect Dis. 2011;15(11):e759-63. [PubMed ID: 21840742]. https://doi.org/10.1016/j.ijid.2011.06.006.
-
17.
Arslankoylu AE, Kuyucu N, Yilmaz BS, Erdogan S. Symptomatic and asymptomatic candidiasis in a pediatric intensive care unit. Ital J Pediatr. 2011;37:56. [PubMed ID: 22104492]. https://doi.org/10.1186/1824-7288-37-56.
-
18.
Malani A, Hmoud J, Chiu L, Carver PL, Bielaczyc A, Kauffman CA. Candida glabrata fungemia: experience in a tertiary care center. Clin Infect Dis. 2005;41(7):975-81. [PubMed ID: 16142662]. https://doi.org/10.1086/432939.
-
19.
Labbe AC, Pepin J, Patino C, Castonguay S, Restieri C, Laverdiere M. A single-centre 10-year experience with Candida bloodstream infections. Can J Infect Dis Med Microbiol. 2009;20(2):45-50. [PubMed ID: 20514159].