Abstract
Introduction:
The purinergic P2X7-receptors and tumor necrosis factor-alpha (TNF-α) may play important roles in the development of pulmonary tuberculosis (PTB). Genetic contribution of the host is among the most important factors that plays a significant role in the susceptibility of the disease. In this regard, both genes for P2X7 receptor and TNF-α have been identified as essential components of the host immune response in the containment of TB. However, the relationship between P2X7 and TNF-α polymorphism and TB susceptibility remains inconclusive.Objectives:
This study was designed to investigate the association of P2X7 and TNF- α gene polymorphisms among Iranian PTB patients.Materials and Methods:
In a case-control study, single nucleotide polymorphisms (SNPs) in P2X7 (+1513, -762) and TNF-α (at -238, -308, -244, -857 and -863) genes were assessed using PCR-RFLP and allele-specific PCR. Thereafter, haplotype and diplotype variability were compared and analyzed.Results:
For the 1513 loci, the heterozygosity was higher in patients (35; 44.3%) than control subjects (12; 24%) [(P = 0.026) ORS; 2.45 CI95 % (1.13 - 5.33)]. For the -762 loci, the frequency of mutant alleles between patients and controls were not statistically significant. No statistical difference was observed in allele frequencies of TNF -308 and -857. However, the frequency of -238 A allele was more in tuberculosis (TB) cases (72.1%) (P = 0.000) [ORs: 5.85 (2.70 - 12.64)]. Data analysis showed greater frequency of haplotypes, i.e. TGGA-CA and CGGA-TA in the patient (21.5%; 14.6%) than control group (2.0%; 6.0%), respectively. Additionally, the diplotype "CCGGGGGGCCAA" was significantly associated with susceptibility to PTB [1.9 (0.08 - 48.3)].Conclusions:
In the studied population, polymorphisms in P2X7 (1513) and TNF-α (S-238) gene were associated with risk of developing PTB. Additionally, distribution of haplotype and diplotype variables did appear to be more specific than SNPs.Keywords
Receptors Purinergic P2X7 Tumor Necrosis Factor-alpha Polymorphism Genetic Tuberculosis
1. Background
Mycobacterium tuberculosis (M. tuberculosis) is an intracellular pathogen that has developed several mechanisms to survive and multiply inside macrophages (1). Generally, the interaction between macrophages and M. tuberculosis has given rise to a cytokine profile that is dominated by tumor necrosis factor-alpha (TNF-α) dependent or independent pathways (2, 3). TNF-α is an important inflammatory mediator and its production is controlled by a gene located in the class III of the MHC region (4). TNF-α is present at sites of active M. tuberculosis infection in humans, regardless of the stage of mycobacterial infection (5). Recently, researchers have shown that extracellular adenosine triphosphate (ATP) induce macrophage bactericidal activity through TNF-α independent pathway, i.e. activation of cell surface P2X7 receptor (5, 6). P2X7 receptor is encoded by the P2RX7 gene (purinergic receptor P2X, ligand-gated ion channel 7 and belongs to P2X receptor family (7, 8)). P2X7 receptor is present on the cells of immune and hempoietic systems. It has been reported that pro-inflammatory cytokines like IL-2, IL-6 and TNF- α up regulate the expression of P2X7 receptor (9, 10), but to date no apparent correlation between the level of P2X7 receptor expression and clinical parameters has been documented. Both P2RX7 and TNF-α genes are polymorphic and several single nucleotide polymorphisms (SNPs) have been identified within them (11-15). Of these, few have been shown to be associated with susceptibility to tuberculosis. For example, SNP at 1513 A to C (rs3751143) abolishes the P2X7 mediated apoptosis of infected macrophages and favors the growth of pathogen (15). Another SNP at the -762 promoter region (T to C) has been shown to be protective against tuberculosis in the Gambian population (16). Similarly, the polymorphisms in the TNF-α gene have been associated with susceptibility to tuberculosis in different ethnic groups (9, 17, 18), but the results have been inconclusive. The aim of this study was to investigate the correlation of polymorphisms in P2X7 genes (1513 and -762) and TNF-α (at -238, -308, -244, -857, -863) in Iranian pulmonary tuberculosis (PTB) patients. Furthermore, the combined polymorphisms (haplotype/diplotype) of these genes were assessed and compared.
2. Objectives
This study was designed to investigate the association of P2X7 and TNF- α gene polymorphisms among Iranian PTB patients.
3. Materials and Methods
3.1. Data Collection
The study included 80 smear and culture positive TB patients that were referred to the Iranian National Reference TB Laboratory from December 2011 to December 2012 for diagnosis and treatment. Additionally, fifty healthy individuals were studied as controls. Control subject were selected from TB personal (clinical and laboratory) who had positive tuberculin tests (10 - 15 mm) and showed no sign of disease (laboratory, radiological and clinical parameter). The Institutional Review Board at the National Research Institute of Tuberculosis and Lung Diseases (NRITLD) approved the study and all the patients signed an informed consent. Patient and control subjects were matched for age, gender, race and nationality.
3.2. DNA Isolation
Genomic DNA was extracted using the standard protocol with slight modifications (12, 19). Briefly, peripheral blood leukocytes (PBLs) were separated from two milliliters of the whole blood using RBC lysis buffer (0.155 M NH4Cl, 0.01 M NaHCO3). Thereafter, PBLs was re-suspended in 500 µL of SE buffer (NaCl 3M, EDTA 0.5M, PH = 8), containing 40 µL of 10% SDS and 3 µL of 20 mg/mL of proteinase K. The suspension was incubated at 600°C for 30 minutes. After incubation, 200 µL of equilibrated phenol (PH = 8) was added to the mixture and centrifuged for 10 minutes at 12000 g. The aqueous phase was transferred to a new tube and the DNA was precipitated using cold propanol (19).
3.3. TNF-α Genotyping
TNF-α gene polymorphisms were studied using PCR- RFLP. For TNF –308 polymorphisms, the following primers were used to amplify a 107 bp product: 5' AGC AAT AGG TGG TTT TGA CTC GGGC CCAT-3'; 5'TCC TCC CTG CTC CGA TTC CG-3'. For -238 and -244 polymorphisms, the following primers were used to amplify a 230 bp product: 5'CCT CAA GGA CTC CAA AGC TTT CTG -3'; 5'ACA CTC CCC ATC CTC CCA GATC -3'. For -857 polymorphisms, the following primers were used to amplify a 127 bp product: 5' GGC TCT GAG GAA TGG GTT AC-3'; 5'CCT CTA CAT GGC CCT GTC TAC-3'. The amplification was accomplished by an initial denaturation at 94°C for 5 minutes, and 30 cycles at 94°C for 40 seconds, at 56°C for 40 seconds, at 72°C for 1 minute, followed by an extension at 72°C for 6 minutes (20, 21).
3.4. PCR-RFLP of TNF-α
PCR products of TNF -238, -244, TNF -308 and TNF -857 were digested with 2 U enzymes of BgI II, Bsaj I, NcoI, TaiI and TaiI, respectively. Digested products were run on 8% polyacrylamide gel, and were stained with silver-nitrate (12, 20-22).
3.5. P2X7 Genotyping
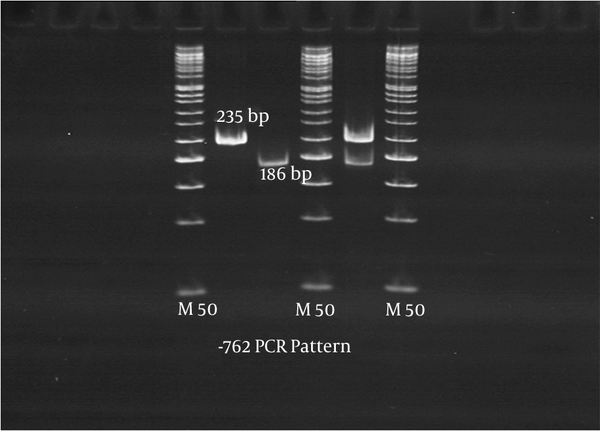
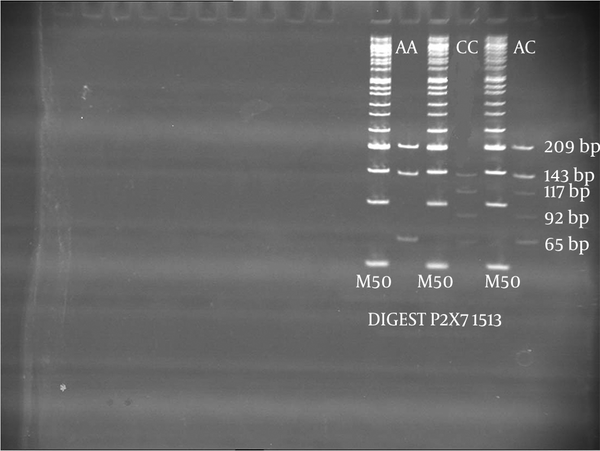
P2X7 gene polymorphism was studied using allele-specific PCR and PCR- RFLP. For P2X7 gene polymorphisms at -762, two outer primers [P2X73 (5'-GAA ACAGGGCCCTGGGTCCTC-3', forward) and P2X74 (5'-TGGTGGGGGTGGAGGGGC- 3', reverse)] and two inner primers [P2X75 (5'-GGTGTCCCTCACTGAATAGGTCAAT-3', forward and P2X76 (5'-GGCAGTCCAACAAAGTTAGGTTTG-3', reverse)] were used. For the -762°C allele, a 235 bp fragment was amplified using the outer forward (P2X73) and inner reverse (P2X76) primers. For the -762 T allele, a 186 bp fragment was amplified using the inner forward (P2X75) and outer reverse (P2X74) primers (6, 18). The amplification was accomplished by an initial denaturation at 94°C for 5 minutes and 30 cycles at 95°C for 30 seconds, at 65°C for 30 seconds, at 72°C for 45 seconds, followed by an extension at 72°C for 10 minutes. The amplified PCR products for CC, CT and TT had the following sizes 235 bp, 186+235 bp and 186 bp, respectively (Figure 1). For P2X7 gene polymorphisms at 1513, the primers 5′ACTCCTAGATCCAGGGATAGCC3′ and 5′TACAGACGTGA GCCACGGT 3′ were used to amplify the 417 bp product (6, 18). The PCR product was digested with 4U enzyme of Hae II. The digested PCR products were run on 8% polyacrylamide gel, which was stained with ethidium bromide. The digest pattern for AA, AC and CC are as follows; 209 + 143 + 65 bp; 209 + 143 + 117 + 92 + 65 bp and 143 + 117 + 95 + 65 bp, respectively (Figure 2).
The -762 Amplified PCR Products
Digest Pattern of 1513 P2X7 Gene
3.6. Statistical Analysis
The frequency of the genotypes in patients and control groups were estimated by direct gene counting and then the data was analyzed using SPSS version 11 (SPSS Inc., Chicago, IL, USA). The odds ratio and P values were calculated for each allele in patient and control groups. All P values were two tailed. A P value of less than < 0.05 was considered significant with 95% confidence interval (CI).
4. Results
4.1. Allele Frequencies of P2X7 Gene Polymorphisms
Each of -762 and 1513 P2X7 polymorphisms showed three types of patterns; frequent homozygote allele (wild type), infrequent heterozygote and infrequent homozygote alleles (mutant type). For the 1513 loci, the heterozygosity (A/C) was higher in patients (35; 44.3%) than controls: (12; 24%) and the difference was statistically significant [(P = 0.026) ORS; 2.45 CI95 % (1.13 - 5.33)]. In contrast, the frequency of wild type (A/A; 74.0% versus 53.1%) was significantly higher in control cases [(P = 0.026) ORS; 0.40 CI95% (0.19 - 0.87)]. However, in both studied groups, the frequency of A allele in 1513 gene polymorphisms was higher than its corresponding mutant types. For the -762 loci, 94.0% of patients had heterozygote mutant alleles (T/C variants) and 6.0% had infrequent homozygote alleles (C/C). No individual carried the wild type (T/T) at -762 in the control group. In general, the frequency of T/C and C/C mutant alleles between patients and controls was not statistically significant. Among patients, two cases (2.5%) showed T/T variables (Table 1).
| Genotypes | Patients, No (%), PTB = 79 | Control Group, No (%), PTB = 50 | Lower Confidence Limit, 95% | Higher Confidence Limit, 95% | OR (95% CI) | Random Effects Weight, % | P Value |
|---|---|---|---|---|---|---|---|
| P2X7-1513 | 42 (53.1) | 37 (74.0) | 0.1905 | 0.8737 | 0.4080 | 7.868 | |
| A/A (frequent type) | 35 (44.3) | 12 (24.0) | 1.1313 | 5.3368 | 2.4571 | 7.8360 | 0.026 |
| A/C (heterozygote mutant) | 2 (2.5) | 1 (2.0) | 0.1363 | 8.3139 | 1.0645 | 4.6739 | 0.024 |
| C/C (homozygote mutant) | NS | ||||||
| P2X7-762 | 2 (2.5) | 0 (0.0) | 0.1532 | 69.277 | 3.258 | 2.9764 | |
| T/T (frequent type) | 67 (84.8) | 47 (94.0) | 0.1149 | 1.3778 | 0.3979 | 6.6051 | NS |
| T/C (heterozygote mutant) | 10 (12.6) | 3 (6.0) | 0.5788 | 7.2627 | 2.0504 | 6.6051 | NS |
| C/C (homozygote mutant | NS | ||||||
| TNF-238 | 57 (72.1) | 2.7088 | 12.6468 | 5.8530 | |||
| A/A (frequent type) | 13 (16.4) | 15 (30.0) | 0.9718 | 15.9600 | 3.9383 | 7.8477 | 0.000 |
| A/G (heterozygote mutant) | 9 (11.3) | 2 (4.0) | 0.0289 | 0.1715 | 0.0704 | 6.2523 | NS |
| G/G (homozygote mutant) | 33 (66.0) | 7.5656 | 0.000 | ||||
| TNF-308 | 1 (1.2) | 0 (0.0) | 0.0771 | 48.3064 | 2.7749 | ||
| A/A (frequent type) | 13 (16.4) | 6 (12.0) | 0.5063 | 3.8150 | 1.9299 | 7.2686 | NS |
| G/A (heterozygote mutant) | 65 (82.2) | 44 (88.0) | 0.2426 | 1.7944 | 1.3898 | 7.2923 | NS |
| G/G (homozygote mutant) | 0.6598 | NS | |||||
| TNF-244 | 79 (100) | 50 (100) | 0.0307 | 80.6038 | 1.5743 | 2.0729 | NS |
| G/G (frequent type) | |||||||
| TNF-857 | 0.3803 | 1.5590 | 0.7700 | 7.9912 | |||
| C/C (frequent type) | 39 (49.3) | 28 (56.0) | 0.8866 | 3.8391 | 1.8449 | 7.9317 | NS |
| C/T (heterozygote mutant) | 37 (46.8) | 16 (32.0) | 0.0811 | 1.2094 | 0.3132 | 6.3787 | NS |
| T/T (homozygote mutant) | 3 (3.7) | 6 (12.0) | NS |
4.2. Allele Frequencies of TNF-α Gene Polymorphisms
The results presented in Table 1, show four types of polymorphisms in the TNF-α gene, an A to G substitution at position -238, a G to A substitution at position -308, a G to A substitution at position-244, and a C to T substitution at position -857. No statistical difference was observed in the allele frequencies of TNF -308 G/A and TNF -857 C/T in control and TB cases. The TNF-α -857 T allele was greater in TB controls (50.5%) than patients (44%) but the differences were not statistically significant. The homozygote mutant alleles for TNF -238 (G/G) were significantly higher in control cases (66.0%) than patients (11.3%) (P = 0.000) [ORs: 0.07 (0.02 - 0.17). In contrast, the frequency of -238 A allele was higher in TB cases (72.1%) than control subjects (30%) (P = 0.000) [ORs: 5.85 (2.70 - 12.64). No individual was found with mutant allele at TNF-α 244 positions.
4.3. Haplotypes Analysis of P2X7 and TNF-α Gene Polymorphisms
In both groups, the most frequent haplotypes of P2X7 genes (-762 & 1513) was CA (50.4%) and TA (29.1%). The other haplotypes CC and TC were greater in patients (5.7%: 19.0%) than controls (1.0%: 13.0%), but the differences was not statistically significant. For four TNF-α gene polymorphisms, seven haplotypes were identified. Out of which the CGGA haplotypes were more frequently seen in patients (48.7%) than controls (28.0%) and the differences was statistically significant (P = 0.001). In comparison the frequency of TGGG was higher in the controls (25%) than patients (4.4%) (Table 2). The combination of TNF-α and P2X7 gene polymorphism haplotypes gives 22 variable forms. The frequencies of TGGA-CA, and CGGA-TA were significantly more in patients (21.5%; 14.6%) than controls (2.0%; 6.0%) [ORs: 10.9 (2.94 - 4.04)]; [ORs: 2.5 (1.02 - 6.25)]. In comparison the haplotype frequencies of CGGG-TA, CGGG-CA, and TGGG-CA were more often seen in control subjects. The haplotype CGGA-CA (18.2%) was the most frequent among both studied groups (Table 2).
| Gene Name | Haplotypes | TB Cases | Control Group | Odds Ratio | P Value | Total Population |
|---|---|---|---|---|---|---|
| P2X7 (-762, 1513) | ||||||
| CA | 78 (49.4) | 52 (52.0%) | 0.9009 [(0.5469 - 1.4838)] | NS | 130 (50.4) | |
| CC | 9 (5.7) | 1 (1.0) | 4.2152 [(0.7394 - 24.o293)] | NS | 10 (3.9) | |
| TA | 41 (25.9) | 34 (34.0) | 0.6808 [(0.3957 - 1.1712)] | NS | 75 (29.1) | |
| TC | 30 (19.0) | 13 (13.0) | 1.5384 [(0.7671 – 3.0853)] | NS | 43 (16.7) | |
| TNF-α (857, 308, 244, 238) | ||||||
| CAGA | 14 (8.9) | 2 (2.0) | 3.9536 [1.0083- 15.5033)] | S0.032 | 16 (6.2) | |
| CAGG | 1 (0.6) | 3 (3.0) | 0.2653 [(0.0385 – 1.8266)] | NS | 4 (1.6) | |
| CGGA | 77 (48.7) | 28 (28.0) | 2.4190 [(1.4191 – 4.1236)] | S0.001 | 105 (40.7) | |
| CGGG | 23 (14.6) | 39 (39.0) | 0.2700 [(0.1492 – 0.4885)] | S 0.000 | 62 (24.0) | |
| TAGG | 0 (0.0) | 1 (1.0) | 0.2093 [(0.0084 – 5.1872)] | NS | 1 (0.4) | |
| TGGA | 36 (22.8) | 2 (2.0) | 11.7397 [(3.1723- 43.4440)] | S0.000 | 38 (14.7) | |
| TGGG | 7 (4.4) | 25 (25.0) | 0.1466 [(0.0620- 0.3463)] | S0.000 | 32 (12.4) | |
| P2X7 & TNF-α | ||||||
| CAGACA | 1 (0.6) | 0 (0.0) | 1.9143 [(0.0772- 47.4520)] | NS | 1 (0.4) | |
| CAGACC | 1 (0.6) | 0 (0.0) | 1.9143 [(0.0772- 47.4520)] | NS | 1 (0.4) | |
| CAGATA | 7 (4.4) | 1 (1.0) | 3.2838 [(0.5587- 19.3019)] | NS | 8 (3.1) | |
| CAGATC | 5 (3.2) | 1 (1.0) | 2.3768 [(0.3840 – 14.7120)] | NS | 6 (2.3) | |
| CAGGTA | 1 (0.6) | 0 (0.0) | 1.9143 [(0.0772 -47.4520)] | NS | 1 (0.4) | |
| CAGGTC | 0 (0.0) | 3 (3.0) | 0.0879 [(0.0045 – 1.7196)] | NS | 3 (1.2) | |
| CGGACA | 32 (20.3) | 15 (15.0) | 1.4172 [(0.7293 – 2.7539)] | NS | 47 (18.2) | |
| CGGACC | 5 (3.2) | 0 (0.0) | 7.2020 [(0.3939 – 131.672)] | NS | 5 (1.9) | |
| CGGATA | 23 (14.6) | 6 (6.0) | 2.5214 [(1.0175- 6.2481)] | S 0.001 | 29 (11.2) | |
| CGGATC | 17 (10.8) | 7 (7.0) | 1.5418 [(0.6304- 3.7710)] | NS | 24 (9.3) | |
| CGGGCA | 6 (3.8) | 16 (16.0) | 0.2183 [(0.0848 – 0.5620)] | S 0.001 | 22 (8.5) | |
| CGGGCC | 2 (1.3) | 0 (0.0) | 3.2109 [(0.1526- 67.5739)] | NS | 2 (0.8) | |
| CGGGTA | 8 (5.1) | 22 (22.0) | 0.1970 [(0.0855 – 0.4541)] | S 0.000 | 30 (11.6) | |
| CGGGTC | 7 (4.4) | 1 (1.0) | 3.2838 [(0.5587 – 19.301)] | NS | 8 (3.1) | |
| P2X7 & TNF-α | ||||||
| TAGGTA | 0 (0.0) | 1 (1.0) | 0.2093 [(0.0084 – 5.1872)] | NS | 1 (0.4) | |
| TGGACA | 34 (21.5) | 2 (2.0) | 10.9181 [(2.9440- 404903)] | S 0.000 | 36 (14.0) | |
| TGGACC | 1 (0.6) | 0 (0.0) | 1.9143 [(0.0772- 47.4520)] | NS | 1 (0.4) | |
| TGGATA | 1 (0.6) | 0 (0.0) | 1.9143 [(0.0772- 47.4520)] | NS | 1 (0.4) | |
| TGGGCA | 5 (3.2) | 19 (19.0) | 0.1498 [0.0560- 0.4005)] | S 0.000 | 24 | |
| TGGGCC | 0 (0.0%) | 1 (1.0%) | 0.2093 [(0.0084-5.1872)] | NS | 1 (0.4%) | |
| TGGGTA | 1 (0.6%) | 4 (4.0%) | 0.2043 [(0.0316 – 1.3187)] | NS | 5 (1.9%) | |
| TGGGTC | 1 (0.6%) | 1 (1.0%) | 0.6317 [(0.0648 – 6.1582)] | NS | 2 |
4.4. Diplotype Analysis of P2X7 and TNF-α Gene Polymorphisms
Out of seven P2X7 diplotypes, the CTAA & CTAC were the most frequent in both studied groups (Table 3). Two diplotypes CCAC (7.6%) and TTAC (2.5%) were seen in patients only. For TNF- α, fifteen diplotypes were detected. The most frequent diplotypes in control groups was CCGGGGAA (24%), CCGGGGGG (24%) and TCGGGGGG (24%). The last two diplotypes were found in very low frequencies among patients (Table 3). The differences were statistically significant. One diplotype, TCGGGGAA, was seen more in patients (27.8%) than control subjects (2.0%) [ORs: 12.9 (2.36 - 7.03)]. As shown in Table 3, combination of TNF-α and P2X7, resulted in 34 variants. The CCGGGGAA-CTAA is the dominant type (14.0%) among studied groups. The diplotype CCGGGGGG-CCAA was significantly associated with susceptibility to PTB [1.9 (0.08 - 48.3)]. In contrast, the diplotype TCGGGGGGCTAA was seen more in controls than patients and the differences was statistically significant (P = 0.003).
| Gene Name | Diplotypes | TB Case | Control Group | Odds Ratio | P Value | Total population |
|---|---|---|---|---|---|---|
| P2X7 (-762, 1513) | ||||||
| CCAA | 3 (3.8) | 3 (6.0) | 0.620 [(0.1351- 2.8537)] | NS | 6 (4.7) | |
| CCAC | 6 (7.6) | 0 (0.0) | 8.932 [(0.4921- 162.11)] | NS | 6 (4.7) | |
| CCCC | 1 (1.3) | 0 (0.0) | 1.9299 [(0.0771- 48.30)] | NS | 1 (0/8) | |
| CTAA | 39 (49.4) | 34 (68) | 0.4665 [(0.2243- 0.9702)] | S 0.046 | 39 (30.5) | |
| CTAC | 27 (34.2) | 12 (24) | 1.6133 [(0.7341-3.5455)] | NS | 73 (56.0) | |
| CTCC | 1 (1.3) | 1 (2) | 0.6306 [(0.0638 – 6.2328)] | NS | 2 (1.5) | |
| TTAC | 2 (2.5) | 0 (0.0) | 3.2581 [(0.1532- 69.2771)] | NS | 2 (1.5) | |
| TNF-α (857, 308, 244, 238) | ||||||
| CCAAGGAA | 1 (1.3) | 0 (0.0) | 1.929 [(0.0771- 48.3064)] | NS | 1 (0.8) | |
| CCGAGGAA | 8 (10.1) | 1 (2.0) | 3.9231 [(0.6661-23.105)] | NS | 9 (7.0) | |
| CCGAGGGG | 0 (0.0) | 1 (2.0) | 0.2075 [(0.0083-5.1957)] | NS | 1 (0.8) | |
| CCGGGGAA | 21 (26.6) | 12 (24.0) | 1.1320 [(0.5050 – 2.5372)] | NS | 33 (25.6) | |
| CCGGGGAG | 5 (6.3) | 2 (4.0) | 1.4322 [(0.3076- 6.6681)] | NS | 7 (5.4) | |
| CCGGGGGG | 4 (5.1) | 12 (24.0) | 0.1836 [(0.0584 – 0.5770)] | S0.002 | 16 (12.4%) | |
| TCGAGGAA | 4 (5.1) | 1 (2.0) | 1.9669 [(0.2994-12.9199)] | NS | 5 (3.9) | |
| TCGAGGGG | 1 (1.3) | 2 (4.0) | 0.3707 [(0.0475-2.8959)] | NS | 3 (2.3) | |
| TCGGGGAA | 22 (27.8) | 22 (27.8) | 12.913 [(2.3687- 70.396)] | S0.001 | 23(17.8%) | |
| TCGGGGAG | 7 (8.9) | 0 (0.0) | 10.448 [(0.5835 – 187.09)] | NS | 7 (5.4) | |
| TCGGGGGG | 3 (3.8) | 12 (24.0) | 0.1409 [(0.0405 – 0.4903)] | S0.001 | 15(11.6%) | |
| TTGAGGGG | 0 (0.0) | 1 (2.0) | 0.2075 [(0.0083 – 5.1957)] | NS | 1 (0.8) | |
| TTGGGGAA | 1 (1.3) | 0 (0.0) | 1.9299 [(0.0771- 48.3064) | NS | 1 (0.8) | |
| TTGGGGAG | 1 (1.3) | 0 (0.0) | 1.9299 [(0.0771- 48.306)] | NS | 1 (0.8) | |
| TTGGGGGG | 1 (1.3) | 5 (10.0) | 0.1581 [(0.0251 – 0.9973)] | S0.032 | 6 (4.7) | |
| P2X7 & TNF-α | ||||||
| CCGAGGAA-CTAA | 1 (1.3) | 0 (0.0) | 1.9299 [(0.0771 – 48.306)] | NS | 1 (0.8) | |
| CCGAGGAA-CTAC | 5 (6.3) | 0 (0.0) | 7.4564 [(0.4034 – 137.839)] | NS | 5 (3.9) | |
| CCGAGGGG-CTAC | 3 (3.8) | 1 (2.0) | 1.5098 [(0.2157 – 10.5680)] | NS | 4 (3.1) | |
| CCGGGGGA-CCCC | 0 (0.0) | 1 (2.0) | 0.2075 [(0.0083 – 5.1957)] | NS | 1 (0.8) | |
| CCGGGGAA-CTAA | 1 (1.3) | 0 (0.0) | 1.9299 [(0.0771- 48.306)] | NS | 1 (0.8) | |
| CCGGGGAA-CTAC | 13 (16.5) | 5 (10.0) | 1.6794 [(0.5814- 4.8511)] | NS | 18 (14.0) | |
| CCGGGGAA-TTAC | 5 (6.3) | 7 (14.0) | 0.4282 [(0.1339- 1.3688)] | NS | 12 (9.3) | |
| CCGGGGAG-CTAA | 2 (2.5) | 0 (0.0) | 3.2581 [(0.1532- 69.2771)] | NS | 2 (1.6) | |
| CCGGGGAG-CTAC | 2 (2.5) | 2 (4.0) | 0.6258 [(0.1045- 3.740)] | NS | 4 (3.1) | |
| CCGGGGGG-CCAA | 3 (3.8) | 0 (0.0) | 4.6209 [(0.2337- 91.3787)] | NS | 3 (2.3) | |
| CCGGGGGG-CTAA | 1 (1.3) | 0 (0.0) | 1.9299 [(0.0771 – 48.306)] | NS | 1 (0.8) | |
| CCGGGGGG-CTAC | 1 (1.3) | 11 (22.0) | 0.0656[ (0.0115- 0.3754)] | S0.000 | 12 (9.3) | |
| TCGAGGAA-CCAC | 2 (2.5) | 1 (2.0) | 1.0645 [(0.1363 – 8.3139)] | NS | 3 (2.3) | |
| TCGAGGAA-CTAA | 1 (1.3) | 0 (0.0) | 1.9299 [(0.0771- 48.306)] | NS | 1 (0.8) | |
| TCGAGGAA-CTAC | 2 (2.5) | 1 (2.0) | 1.0645 [(0.1363- 8.3139)] | NS | 3 (2.3) | |
| TCGAGGGG-CTAA | 1 (1.3) | 0 (0.0) | 1.0299 [(0.0771- 48.306)] | NS | 1 (0.8) | |
| TCGAGGGG-CTAC | 1 (1.3) | 0 (0.0) | 1.0299[(0.0771 – 48.3064)] | NS | 1 (0.8) | |
| TCGGGGAA-CCAA | 0 (0.0) | 2 (4.0) | 0.1222 [(0.0057- 2.5952)] | NS | 2 (1.6) | |
| TCGGGGAA-CCAC | 1 (1.3) | 0 (0.0) | 1.9299 [(0.0771 – 48.3064)] | NS | 1 (0.8) | |
| TCGGGGAA-CTAA | 3 (3.8) | 0 (0.0) | 4.6209 [(0.2337 – 91.3787)] | NS | 3 (2.3) | |
| TCGGGGAA-CTAC | 8 (10.1) | 1 (2.0) | 3.9231 [(0.6661 – 23.1056)] | NS | 9 (7.0) | |
| TCGGGGAA-CTCC | 9 (11.4) | 0 (0.0) | 13.609 [(0.7742- 239.242)] | NS | 9 (7.0) | |
| TCGGGGAG-CCAA | 1 (1.3) | 0 (0.0) | 1.9299 [(0.0771- 48.3064)] | NS | 1 (0.8) | |
| TCGGGGAG-CCAC | 1 (1.3) | 0 (0.0) | 1.9299 [(0.0771- 48.3064)] | NS | 1 (0.8) | |
| TCGGGGAG-CTAA | 1 (1.3) | 0 (0.0) | 1.9299 [(0.0771- 48.3064)] | NS | 1 (0.8) | |
| TCGGGGAG-CTAC | 3 (3.8) | 0 (0.0) | 4.6209 [(0.2337 – 91.3787)] | NS | 3 (2.3) | |
| TCGGGGGG-CCAA | 2 (2.5) | 0 (0.0) | 3.2581 [(0.1532 – 69.2771)] | NS | 2 (1.6) | |
| TCGGGGGG-CCAC | 0 (0.0) | 3 (6.0) | 0.0854 [(0.0043 – 1.6887)] | NS | 3 (2.3) | |
| TCGGGGGG-CTAA | 1 (1.3) | 0 (0.0) | 1.9299 [(0.0771 – 48.3064)] | NS | 1 (0.8) | |
| TTGAGGGG-CTAA | 2 (2.5) | 9 (18.0) | 0.1409 [(0.0333 – 0.5967)] | S0.003 | 11 (8.5) | |
| TTGGGGAA-CTAA | 0 (0.0) | 1 (2.0) | 0.2075 [(0.0083 – 5.1957)] | NS | 1 (0.8) | |
| TTGGGGAG-CTAC | 1 (1.3) | 0 (0.0) | 1.9299 [(0.0771 – 48.3064)] | NS | 1 (0.8) | |
| TTGGGGGG-CTAA | 1 (1.3) | 0 (0.0) | 1.9299 [(0.0771-48.3064)] | NS | 1 (0.8) | |
| TTGGGGGG-CTCC | 1 (1.3) | 4 (8.0) | 0.1975 [(0.0300 - 1.2981)] | NS | 5 (3.9) | |
| TTGGGGGG-CTCC | 0 (0.0) | 1 (2.0) | 0.2075 [(0.0083 - 5.1957)] | NS | 1 (0.8) |
5. Discussion
This study was conducted to assess the rate of human P2X7 and TNF-α gene polymorphisms among Iranian pulmonary TB cases. Genetic variability and environmental factors usually contributed to the risk of developing active tuberculosis (12, 23, 24). For instance, the possible association between polymorphisms in P2X7 and susceptibility to TB has gained considerable attention, during the recent years (6, 11, 14). P2X7 gene is highly polymorphic, and it is up-regulated by an inflammatory cytokine (9, 10). Activation of P2X7 receptors induces permeabilization or apoptosis in cells of immune systems (3, 11, 24-26). Wiley and coworkers identified four losses of function in single amino acid substitutions (27). In 2002 Li et al. outlined the ethnic and racial differences in P2X7, later on, Ferrari et al. identified the first gain of P2X7 functional polymorphism (3, 7, 24). Based on these observations, a link between P2X7 polymorphisms and reactivation of certain intracellular pathogens particularly MTB were proposed. We found a higher frequency of 1513 C allele in patients (46.8%) than control (26%) subject. Therefore, a higher risk of developing tuberculosis in individuals with hetero or homozygous allele of 1513 (AC; CC) was highlighted. Some studies documented the importance of the 1513 C allele with extra-pulmonary TB (27). However, the mechanisms by which these genotypes can protect or support the pathogencity of TB have not been completely elucidated. Yet, a few investigators showed an association between 1513 C allele and loss of P2X7 receptor function which could protect the host from pathogen by eliminating the MTB infected macrophages (3, 6, 18, 27). Additional to 1513 polymorphism, -762 T to C allele frequency in the promoter region, can influence the degree to which the receptor is down regulated via other host or pathogen-generated inhibitory factors. In our study, -762 variant of P2X7 was not associated with PTB among Iranian studied cases. In contrast to our study, Bahari et al. found a significant association between -762 TC and tuberculosis (28). Overall, six main ethnic groups live in Iran; Fars, Turk, Kord, Lor, Arab, and Sistanei. In our study, patients were selected from the "Fars" ethnic group, whereas Bahari studied Sistanei patients. Additionally, the control subjects in our study were selected from individuals who were working in TB wards or TB laboratories and had had adequate exposure time for the pathogen to cause disease. The time of exposure was from five years (minimum) to 15 years (maximum). Control subjects were screened by laboratory and clinical parameters, but they were and remained negative for tuberculosis. Therefore, we believe that besides ethnic and racial differences, environmental and socio-economic conditions of the host could compromise disease development. Basically, cytokines production is effected by SNP in the promoter or coding regions of cytokine genes that directly affect the transcription and synthesis of mRNA (28, 29). Today, it is known that the distribution and frequency of SNP or polymorphisms are variable among different ethnic groups e.g. the polymorphisms in the promoter region of -762 TC had a significant association with tuberculosis in Gambian and Indian patients, whereas the -762 TC variant was not found to be significantly associated with tuberculosis in the Chinese Han Population. Recently, Zhang et al. outlined the influence of different pro inflammatory cytokines like IL-2, IL-6 and TNF- α in up –regulation of P2X7 synthesis (9). Later on, Franco-Martinez et al. proposed q correlation between elevated levels of TNF- α and expression of P2X7 receptor in pulmonary tuberculosis patients (10), but they could not document any apparent clinical correlation.
As polymorphisms in TNF-α gene may affect TNF-α production, we tried to analysis the SNP at four regions of -238, -308, -244 and -857, followed by comparison of their variability with the P2X7 polymorphisms in the same patient. Our findings showed that the SNP alone may not be associated with PTB, but the frequency of TNF-α haplotypes at 857, 308, 244 and 238 (CAGA, CGGA, TGGA, and TGGG) were significantly higher in patients than control subjects. Similarly, we found one single diplotype (TCGGGGAA) in TNF-α (857, 308, 244, 238), which was significantly associated with susceptibility to PTB (Table 3). The frequency of this diplotype was 27.8% in patients versus 2% in control subjects. Likewise, the assessment of diplotype frequencies in P2X7 gene was more reliable than SNP analysis. Hence, we conclude that the study of single nucleotide polymorphisms is a bit of a traditional approach and for identifying the candidate genes in molecular epidemiology we need to investigate the frequency of haplotype or diplotype polymorphisms (12, 30, 31). For examples the SNP at 857, 308 and 244 of TNF- α gene were not associated with studied TB patients, but when we combined the allele frequencies of P2X7 and TNF-α together, 22 haplotypes and 34 diplotypes were identified. A significant association with PTB susceptibility was seen with two haplotypes namely: TGGA-CA (21.5%) and CGGA-TA (14.6%). The frequencies of these haplotypes in control subjects were very low, 2.0% and 6.0%, respectively (Table 2 and 3). Based on this information, haplotype and/or diplotype variables can be considered as an alternative way to study the SNPs. In conclusion, the present study supports the association between P2X7 and TNF-α and susceptibility to PTB. The frequency of diplotypes in our studied population was more specifically linked to susceptibility to PTB than haplotypes. Further prospective studies of these polymorphisms particularly haplotypes and diplotypes are warranted to identify people who are at high risk of developing TB.
Acknowledgements
References
-
1.
DesJardin LE, Kaufman TM, Potts B, Kutzbach B, Yi H, Schlesinger LS. Mycobacterium tuberculosis-infected human macrophages exhibit enhanced cellular adhesion with increased expression of LFA-1 and ICAM-1 and reduced expression and/or function of complement receptors, FcgammaRII and the mannose receptor. Microbiology. 2002;148(Pt 10):3161-71. [PubMed ID: 12368450].
-
2.
Oddo M, Renno T, Attinger A, Bakker T, MacDonald HR, Meylan PR. Fas ligand-induced apoptosis of infected human macrophages reduces the viability of intracellular Mycobacterium tuberculosis. J Immunol. 1998;160(11):5448-54. [PubMed ID: 9605147].
-
3.
Ferrari D, Los M, Bauer MK, Vandenabeele P, Wesselborg S, Schulze-Osthoff K. P2Z purinoreceptor ligation induces activation of caspases with distinct roles in apoptotic and necrotic alterations of cell death. FEBS Lett. 1999;447(1):71-5. [PubMed ID: 10218585].
-
4.
Schwander SK, Torres M, Carranza CC, Escobedo D, Tary-Lehmann M, Anderson P, et al. Pulmonary mononuclear cell responses to antigens of Mycobacterium tuberculosis in healthy household contacts of patients with active tuberculosis and healthy controls from the community. J Immunol. 2000;165(3):1479-85. [PubMed ID: 10903753].
-
5.
Islam N, Kanost AR, Teixeira L, Johnson J, Hejal R, Aung H, et al. Role of cellular activation and tumor necrosis factor-alpha in the early expression of Mycobacterium tuberculosis 85B mRNA in human alveolar macrophages. J Infect Dis. 2004;190(2):341-51. [PubMed ID: 15216471]. https://doi.org/10.1086/421522.
-
6.
Mokrousov I, Sapozhnikova N, Narvskaya O. Mycobacterium tuberculosis co-existence with humans: making an imprint on the macrophage P2X(7) receptor gene? J Med Microbiol. 2008;57(Pt 5):581-4. [PubMed ID: 18436590]. https://doi.org/10.1099/jmm.0.47455-0.
-
7.
Li CM, Campbell SJ, Kumararatne DS, Bellamy R, Ruwende C, McAdam KP, et al. Association of a polymorphism in the P2X7 gene with tuberculosis in a Gambian population. J Infect Dis. 2002;186(10):1458-62. [PubMed ID: 12404161]. https://doi.org/10.1086/344351.
-
8.
Khakh BS, North RA. P2X receptors as cell-surface ATP sensors in health and disease. Nature. 2006;442(7102):527-32. [PubMed ID: 16885977]. https://doi.org/10.1038/nature04886.
-
9.
Zhang XJ, Zheng GG, Ma XT, Lin YM, Song YH, Wu KF. Effects of various inducers on the expression of P2X7 receptor in human peripheral blood mononuclear cells. Sheng Li Xue Bao. 2005;57(2):193-8. [PubMed ID: 15830104].
-
10.
Franco-Martinez S, Nino-Moreno P, Bernal-Silva S, Baranda L, Rocha-Meza M, Portales-Cervantes L, et al. Expression and function of the purinergic receptor P2X7 in patients with pulmonary tuberculosis. Clin Exp Immunol. 2006;146(2):253-61. [PubMed ID: 17034577]. https://doi.org/10.1111/j.1365-2249.2006.03213.x.
-
11.
Miller CM, Boulter NR, Fuller SJ, Zakrzewski AM, Lees MP, Saunders BM, et al. The role of the P2X(7) receptor in infectious diseases. PLoS Pathog. 2011;7(11). ee1002212. [PubMed ID: 22102807]. https://doi.org/10.1371/journal.ppat.1002212.
-
12.
Merza M, Farnia P, Anoosheh S, Varahram M, Kazampour M, Pajand O, et al. The NRAMPI, VDR and TNF-alpha gene polymorphisms in Iranian tuberculosis patients: the study on host susceptibility. Braz J Infect Dis. 2009;13(4):252-6. [PubMed ID: 20231985].
-
13.
Fan HM, Wang Z, Feng FM, Zhang KL, Yuan JX, Sui H, et al. Association of TNF-alpha-238G/A and 308 G/A gene polymorphisms with pulmonary tuberculosis among patients with coal worker's pneumoconiosis. Biomed Environ Sci. 2010;23(2):137-45. [PubMed ID: 20514989]. https://doi.org/10.1016/S0895-3988(10)60043-8.
-
14.
Gu BJ, Zhang W, Worthington RA, Sluyter R, Dao-Ung P, Petrou S, et al. A Glu-496 to Ala polymorphism leads to loss of function of the human P2X7 receptor. J Biol Chem. 2001;276(14):11135-42. [PubMed ID: 11150303]. https://doi.org/10.1074/jbc.M010353200.
-
15.
Sharma S, Rathored J, Ghosh B, Sharma SK. Genetic polymorphisms in TNF genes and tuberculosis in North Indians. BMC Infect Dis. 2010;10:165. [PubMed ID: 20537163]. https://doi.org/10.1186/1471-2334-10-165.
-
16.
Ben-Selma W, Ben-Kahla I, Boukadida J, Harizi H. Contribution of the P2X7 1513A/C loss-of-function polymorphism to extrapulmonary tuberculosis susceptibility in Tunisian populations. FEMS Immunol Med Microbiol. 2011;63(1):65-72. [PubMed ID: 21635566]. https://doi.org/10.1111/j.1574-695X.2011.00824.x.
-
17.
Wang Q, Zhan P, Qiu LX, Qian Q, Yu LK. TNF-308 gene polymorphism and tuberculosis susceptibility: a meta-analysis involving 18 studies. Mol Biol Rep. 2012;39(4):3393-400. [PubMed ID: 21735105]. https://doi.org/10.1007/s11033-011-1110-x.
-
18.
Sharma S, Kumar V, Khosla R, Kajal N, Sarin B, Sehajpal P. Association of P2X7 receptor +1513 (A-->C) polymorphism with tuberculosis in a Punjabi population. Int J Tuberc Lung Dis. 2010;14(9):1159-63. [PubMed ID: 20819262].
-
19.
Sambrook J, Russell DW. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press; 2001.
-
20.
Yen JH, Chen CJ, Tsai WC, Lin CH, Ou TT, Wu CC, et al. Tumor necrosis factor promoter polymorphisms in patients with rheumatoid arthritis in Taiwan. J Rheumatol. 2001;28(8):1788-92. [PubMed ID: 11508580].
-
21.
Barber MD, Powell JJ, Lynch SF, Gough NJ, Fearon KC, Ross JA. Two polymorphisms of the tumour necrosis factor gene do not influence survival in pancreatic cancer. Clin Exp Immunol. 1999;117(3):425-9. [PubMed ID: 10469042].
-
22.
Anoosheh S, Farnia P, Kargar M. Association between TNF-Alpha (-857) Gene Polymorphism and Susceptibility to Tuberculosis. Iran Red Crescent Med J. 2011;13(4):243-8. [PubMed ID: 22737473].
-
23.
Amirzargar AA, Rezaei N, Jabbari H, Danesh AA, Khosravi F, Hajabdolbaghi M, et al. Cytokine single nucleotide polymorphisms in Iranian patients with pulmonary tuberculosis. Eur Cytokine Netw. 2006;17(2):84-9. [PubMed ID: 16840026].
-
24.
Cooke GS, Hill AV. Genetics of susceptibility to human infectious disease. Nat Rev Genet. 2001;2(12):967-77. [PubMed ID: 11733749]. https://doi.org/10.1038/35103577.
-
25.
Ferrari D, Pizzirani C, Adinolfi E, Lemoli RM, Curti A, Idzko M, et al. The P2X7 receptor: a key player in IL-1 processing and release. J Immunol. 2006;176(7):3877-83. [PubMed ID: 16547218].
-
26.
Saunders BM, Fernando SL, Sluyter R, Britton WJ, Wiley JS. A loss-of-function polymorphism in the human P2X7 receptor abolishes ATP-mediated killing of mycobacteria. J Immunol. 2003;171(10):5442-6. [PubMed ID: 14607949].
-
27.
Wiley JS, Dao-Ung LP, Li C, Shemon AN, Gu BJ, Smart ML, et al. An Ile-568 to Asn polymorphism prevents normal trafficking and function of the human P2X7 receptor. J Biol Chem. 2003;278(19):17108-13. [PubMed ID: 12586825]. https://doi.org/10.1074/jbc.M212759200.
-
28.
Bahari G, Hashemi M, Taheri M, Naderi M, Moazeni-Roodi A, Kouhpayeh HR, et al. Association of P2X7 gene polymorphisms with susceptibility to pulmonary tuberculosis in Zahedan, Southeast Iran. Genet Mol Res. 2013;12(1):160-6. [PubMed ID: 23408402]. https://doi.org/10.4238/2013.January.24.8.
-
29.
Padyukov L, Hahn-Zoric M, Lau YL, Hanson LA. Different allelic frequencies of several cytokine genes in Hong Kong Chinese and Swedish Caucasians. Genes Immun. 2001;2(5):280-3. [PubMed ID: 11528523]. https://doi.org/10.1038/sj.gene.6363771.
-
30.
Skorpil N, Kolesar L, Striz I, Lardy NM, Slavcev A. Cytokine gene polymorphisms in the Dutch population. Int J Immunogenet. 2007;34(2):87-90. [PubMed ID: 17373932]. https://doi.org/10.1111/j.1744-313X.2007.00663.x.
-
31.
Fairfax BP, Davenport EE, Makino S, Hill AV, Vannberg FO, Knight JC. A common haplotype of the TNF receptor 2 gene modulates endotoxin tolerance. J Immunol. 2011;186(5):3058-65. [PubMed ID: 21282507]. https://doi.org/10.4049/jimmunol.1001791.