1. Background
Coagulase-negative Staphylococci (CoNS), and especially Staphylococcus epidermidis, are considered to be commensal bacteria of human skin and oral-nasal mucosa (1). However, this microorganism is implicated in several opportunistic infections such as eye keratitis and endophthalmitis of contaminated contact lenses (2), urinary catheters (3), prosthetic joints, and bloodstream infections particularly in neonatal intensive care units (NICUs) (4). In addition, a wide variety of infections related to foreign bodies such as medical implant devices, peripheral or central intravenous catheters (CVCs), and indwelling catheters are also attributed to S. epidermidis (5). On the other hand, many virulence factors including elastase, lipase, esterase, delta toxin, and biofilm (polysaccharide intercellular adhesion) have been recognized in S. epidermidis (6).
S. epidermidis is known as a major cause of hospital-acquired bacteremia and invasive nosocomial infections (7, 8). Recently, the resistance of S. epidermidis to antimicrobial agents, particularly to methicillin, has been reported in various hospitals around the world (9). The emergence of methicillin-resistant S. epidermidis (MRSE) and also multidrug-resistant (MDR) strains are demonstrated worldwide (10). Moreover, the existence of a variety of adhesion factors and the ability for aggregation as well as biofilm formation are the main reasons for bacterial survival and circulation in the hospital setting.
The MRSE and also MDR strains are regarded as part of the repertoire of antimicrobial resistance genes that can be transferred to other staphylococci species (11, 12). Persistence of multidrug-resistant clones of S. epidermidis in health care units may lead to true infections in high-risk patients. Hence, the assessment of the route of infection and also discrimination between true infection and contamination is of prime importance.
2. Objectives
The aim of this study was to determine the molecular epidemiology of S. epidermidis true infection-associated isolates recovered from admitted patients in a referral hospital of Isfahan..
3. Methods
3.1. Setting
This study was conducted at the Al-Zahra hospital; a main tertiary referral hospital affiliated with Isfahan University of Medical Sciences with 950 beds. This hospital provides tertiary services in different wards, including an intensive care unit (ICU) and nephrology, neonatal, oncology, urology, and neurology wards. The emergency unit of this hospital is the main referral center for patients from different parts of Isfahan and the neighboring provinces. Also, the Department of Pediatrics provides a variety of services for neonates and infants. According to the assessment of the ministry of health in 2010 - 2011, this center achieved the first rank among Iranian hospitals.
During the 8-month study period, several patients were screened for S. epidermidis infection/colonization. Demographic data and history of each case such as age, gender, ward of admission, site, and date of isolation were recorded. All S. epidermidis isolates were classified as either contaminated or true infections with following criteria: microscopic observation of pus cells (WBCs) in the specimens by a trained medical technologist in the hospital laboratory, a threefold isolation of bacteria, and an accepted colony count. Since the surveillance definition of the CDC states that bloodstream infections should be confirmed, necessary requirements such as patients with fever, hypothermia or hypotension were also taken into account (13).
3.2. Identification of Isolates
S. epidermidis isolates were identified using the following tests: colony morphology, Gram staining, catalase, coagulase test with rabbit plasma, DNase activity, mannitol fermentation, acid from trehalose, polymyxin B resistance, and other specific microbiological standard tests. The strains were stored in Trypticase soy broth (TSB) with 10% sterile glycerol at -20°C.
3.3. Antibiotic Susceptibility Testing
An antibiotic susceptibility test was performed using Muller Hinton Agar (MHA) by disk diffusion method with ten antibiotic discs (Mast, Merseyside, England) including: ciprofloxacin (5 µg), gentamicin (10 µg), clindamycin (2 µg), cefoxitin (30 µg), oxacillin (1 µg), tetracycline (30 µg), levofloxacin (5 µg), trimethoprim (1.25 µg) + sulfamethoxazole (23.75 µg), and rifampin (5 µg) according to the CLSI (2013) guidelines (14). Isolates with an intermediate level of susceptibility were classified as non-susceptible. We defined the S. epidermidis isolates as MDR strains by observing the resistance to at least 3 antibiotics of different classes.
3.4. Pulsed Field Gel Electrophoresis (PFGE)
Forty isolates, which were associated with true infections, were characterized by PFGE. PFGE was conducted as described by Chung et al. (15). In brief, one colony from each isolate was inoculated into a 5 mL Brain Heart Infusion Broth (BHI) and incubated for 24 hours at 35 - 37°C. The optical density of each sample test reached 1.2 - 1.8 at 600 nm. Each plug was prepared by adding 120 μL of bacterial suspension, 400 μL EC buffer (10 mM EDTA, 1 M NaCl, 6 mM Tris-HCl (pH 7.6), 0.2% Deoxycholate, 0.5% Sarkosyl and 0.5% Brij-58), 20 μL Lysostaphin (1 mg/mL) (Sigma, USA) and 450 μL 2% Seakem Gold agarose (FMC Bio-Products, Rock land, ME). By adding 1800 μL EC buffer with 200 μL lysozyme (20 mg/mL) (Sigma, USA), plugs were incubated in 37°C for 5 hours, then washed with 1 × TE buffer (10 mM Tris-HCl, 1 mM EDTA). Thereafter, 1800 μL ES buffer (0.4M EDTA, 1% Sarkosyl) plus 70 μL of 50 mg/mL proteinase K (Sigma, USA) were added and incubated at 55°C overnight. The restriction digestion step was done by adding 30 U SmaI (Takara, Japan) for an incubation of 16 - 20 hours at 30°C after washing the plug slices with TE buffer.
PFGE was performed with 1% agarose gel in a contour-clamped homogeneous electric field (CHEF-DRIII) (Bio-Rad USA) apparatus for 22 hours overnight at 12°C, with a constant voltage of 6 V/cm and pulses ramped from 5 to 50 seconds at an angle of 120 degrees. XbaI (Takara, Japan) digested genomic DNA of the Salmonella enterica serovar Braenderup strain H9812, which was used as a molecular weight marker. Gels were photographed after staining in 0.5 µg/mL ethidium bromide. Finally, PFGE profile were analyzed with GelCompar II version 4 (Applied Maths, Sint-Martens-Latem, Belgium) with a 90% similarity cut-off point and clustered using the Dice’s coefficient, then represented by unweighted pair grouping by mathematical averaging (UPGMA). The band pattern of each strain was also visually confirmed according to the principles stated by Tenover et al. (16).
3.5. Multilocus Sequence Typing (MLST)
Nine isolates of distinct pulsotypes were evaluated by MLST. MLST was carried out though comparing the nucleotide sequences of 7 housekeeping genes, consisting of carbamate kinase (arcC), shikimate dehydrogenase (aroE), ABC transporter (gtr), DNA mismatch repair protein (mutS), pyrimidine operon regulatory protein (pyrR), triosephosphate isomerase (tpiA,) and acetyl-CoA acetyltransferase (yqiL), which was previously described by Thomas et al. (17). The alleles, allelic profiles, and the sequence type (STs) of each of the 7 housekeeping loci were obtained from the MLST database (http://www.mlst.net).
3.6. Data Analysis
All statistical analyses were performed with SPSS version 21 (SPSS Inc., Chicago, IL). Each PFGE type was considered as a pulsotype and the combination of allelic types was defined as sequence type (ST).
4. Results
In our cross sectional study among 183 recovered S. epidermidis isolates from different patients, 40 isolates were recognized as true infection-associated strains. The most commonly collected specimen was from the bloodstream (35%) as well as from wounds (35%), followed by catheter related infections (10%), and other sites (20%) including shunt, joint, and surgical sites. Using the antimicrobial susceptibility test, 75% of the tested isolates were found to be resistant to cefoxitin, 60% of isolates to ciprofloxacin, and 45% to tetracycline. Furthermore, resistance to oxacillin was 55%, gentamicin 50%, sulfamethoxazole/trimethoprim 47.5%, clindamycin 47.5%, levofloxacin 32.5% and rifampin 27.5%. More than one third (37.5%) of the isolates were multidrug-resistant.
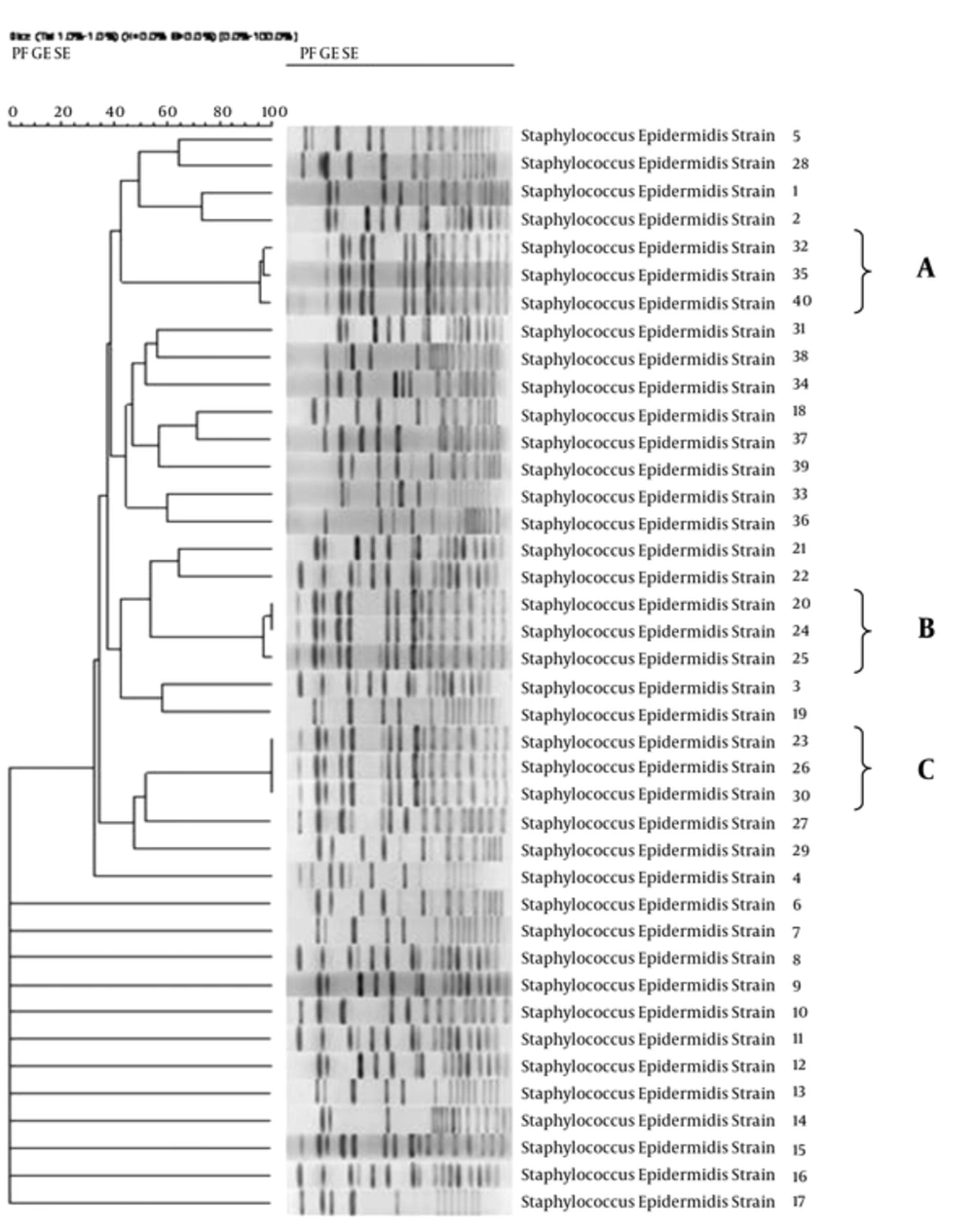
By GelCompar II analysis, a total of 34 different pulsotypes were identified, including 3 clones (A, B, C) and 31 singletons. In each clone, 3 strains were found (Figure 1). Details of the strains are illustrated in Table 1.
| Clone Name | Strain | Ward | Specimen |
|---|---|---|---|
| A | A1 | Surgery | Ear wound |
| A2 | NICU | Bloodstream | |
| A3 | NICU | Bloodstream | |
| B | B1 | NICU | Bloodstream |
| B1 | NICU | Bloodstream | |
| B2 | Oncology | Bloodstream | |
| C | C1 | CCU | Catheter |
| C1 | NICU | Eye wound | |
| C1 | ICU | Bloodstream |
In total, 5 different STs were identified (ST2, ST5, ST27, ST29, and ST66). The 3 strains of clone A (A1, A2, and A3) belonged to ST2, ST27, and ST66; clone B (B1, B1 and B2) belonged to ST2, ST2, and ST20, whereas, the same 3 strains of clone C (C1) were related to ST5.
5. Discussion
The importance of S. epidermidis and related infections among coagulase negative staphylococci has been revealed in many studies (18). Villari et al. have described that 29.8% of surface infections and 39.8% of bloodstream infections belonged to S. epidermidis (19). Recently, as a result of the emergence of multidrug-resistant strains (MDR), the S. epidermidis nosocomial infections have become a major challenge in health care units. In our research, 37.5% of the isolates were determined as MDR with resistance to 3 different classes of antibiotics. In addition, a high rate of resistance to oxacillin (55%) was found in our clinical isolates. Moreover, the majority of the strains were resistant to ciprofloxacin, tetracycline, and trimethoprim/sulfamethoxazole as well as oxacillin. These findings are consistent with other studies (20). Discussions regarding CoNS in blood cultures and hospital infections in numerous European and Asian countries have demonstrated that resistance to aminoglycosides and trimethoprim/sulfamethoxazole has increased in past two decades (20, 21). Increase of MDR strains and the importance of functional molecular typing methods in comparison to conventional techniques has motivated us to conduct the present study. This study, as the first report of S. epidermidis molecular epidemiology from a main tertiary referral hospital in Isfahan, Iran, characterized the 40 true clinical S. epidermidis isolates, using PFGE as a short-term epidemiological method. Our results showed that a total 34 different pulsotypes with 3 clones (A, B, and C) were distinguished. These results indicate that S. epidermidis has a high degree of genotypic diversity, as found in other investigations. For instance, in 2002, Raimundo et al. identified 43 subtypes in 55 isolates in a neonatal intensive care unit, although related studies have shown different clinical outcomes (22). However, we have found that a number of isolates were different in pulsotypes while, displaying a similar antibiotic resistance profile (strains 9 and 10 in this study), the association between the PFGE subtypes and various resistance profiles were equally observed as in the study of Miragaia et al. in 2002 (23). The microorganism gene exchange and common genetic events such as horizontal gene transfer and dissemination of mobile genetic elements may play an important role in pattern diversity (24).
As a result of S. epidermidis infections in different health care units, application of molecular epidemiology typing methods is a helpful technique for tracking the clones and controlling the further transmission of infections. Amongst several methods for molecular typing, pulsed-field gel electrophoresis (PFGE) is the preferential method for S. epidermidis epidemiological studies and outbreaks (16). Furthermore, multilocus sequence typing (MLST) is known as an alternative molecular typing method for evaluating the taxonomic classification and microorganism genetic diversity in long-term epidemiological studies (17).
In the present study, the molecular characteristics of 3 clones (9 strains) in PFGE were examined using the MLST method. The MLST resulted in 5 distinct STs and showed that both ST2 and ST5 were the most represented sequence types. Although these STs have previously been detected in different studies, this is the first report originating from our tertiary referral hospital in Isfahan. Most studies have shown that ST2 is associated with positive isolates of the intercellular adhesion gene (ica), which has been disseminated in many regions and has caused a variety of serious infections. Comparatively, ST27 is highly adapted to hospital environments since it is rarely found outside of medical facilities and can therefore be colonized in hospitalized patients in a short period of time (25, 26).
Considering the main clones, pulsotypes, and identified STs, the dissemination of MDR clones occurred in the hospital setting. These strains were found in different wards of the hospital such as the surgery and oncology wards, the neonatal intensive care unit (NICU), the intensive care unit (ICU), and the coronary care unit (CCU). This issue is more evidently seen in clone C, in which the first strain was isolated from the CCU ward from a catheter sample, the second strain from the NICU (eye wound) and the third from the ICU (bloodstream) (Table 1).
In accordance with the life cycle of S. epidermidis, both our results and the increase of MDR as well as invasive strains implicated in severe diseases, infection control procedures and prevention of colonization are essential for control of the epidemic strains in health care units.