Abstract
Background:
Brucellosis is a zoonotic disease that causes major economic and public health problems. It is one of the most important diseases in humans and domestic animals. Hence, the exact identification of Brucella spp. is important for strategies of treatment and control. Polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) is one of the molecular techniques characterized by amplification of deoxyribonucleic acid (DNA) sequence and restriction enzyme digestion.Objectives:
This study aimed at identifying genetic polymorphisms of omp2a genes among 90 Brucella isolated from humans and animals, using the PCR-RFLP method.Methods:
Ninety Brucella spp. isolated from humans and animals in two different regions of Iran were used in this study. Biochemical tests and the Brucella omp2a (1100 bp) gene-PCR was used for identification of Brucella isolates. Polymerase Chain Reaction products were digested by restriction endonuclease enzyme pstI and gene sequencing analysis was carried out for molecular typing of Brucella strains. Therefore, genetic relatedness was revealed by a dendrogram.Results:
Analysis of the 90 Brucella strains by biochemical tests, PCR, and PCR-RFLP methods with PstI enzyme and omp2a sequencing showed four unique RFLP Profiles (P1-P4). Seventy-nine (87.8%) of the Brucella isolates belonged to B. melitensis strain 20236. From 30 animal isolates, nine (30%) belonged to B. melitensis biovare1 and two (6.6%) to B. abortus strain. According to the RFLP dendrogram, group 1 and 2 had higher genetic relatedness similarity.Conclusions:
The results showed B. melitensis strain 20236 was the predominant strain among human and animal Brucella isolates. Likewise, according to dendrogram results, the PCR-RFLP technique was not able to separate human and animal species of B. melitensis from B. abortus.Keywords
1. Background
Brucella is a Gram-negative facultative intracellular bacillus that can infect many human and animal species. Conventionally, they are classified to seven species, each of them comprised of several biovars (1). Annually, more than 500,000 human cases of brucellosis are reported to the world health organization (2). The prevalence of brucellosis in Iran was reported as 0.5% to 10.9%, in different provinces (3). Incidence rate of brucellosis was about 66 to 100 per 100,000 population in the province of Hamadan, Iran (4). Brucellosis still remains an unrestrained problem in many regions, such as the Middle East, Mediterranean countries, Latin America, Africa, and some parts of Asia (5). The disease is endemic and previous studies have reported that B. melitensis biotype 1 is the predominant strain in human and animal isolates in Iran (1, 6).
Distinction between species and biovars of Brucella spp. is currently based on differential tests, such as phenotypic characterization of lipopolysaccharides (LPS) antigens, dye-sensitivity, H2S production, phage typing, CO2 requirement, and metabolic properties (7). However, phenotypic tests are less sensitive and not always reliable (8). While, some species of Brucella may be characterized by serological tests, they are not able to discriminate between species and biovars (9). Homology of DNA for all species in the genus of Brucella is more than 90% and probable new biovars and strains of Brucella enter the area. Consequently, investigations about the epidemiology of brucellosis, pathogenicity, and ecological differences between species and biovars of Brucella may help choose the correct vaccine, plan epidemiological studies, and control and eradicate the condition (10, 11). Molecular techniques have shown accurate typing of Brucella spp. based on specific identification of Brucella nucleotide sequences associated with the genus, species, and biovars. Therefore, these methods are important tools for diagnosis in epidemiologic studies (12).
Furthermore, PCR-RFLP is a molecular technique applied by amplification of DNA sequence and restriction enzyme digestion. However, RFLP analysis is widely used for the identification of bacterial species and biovars of the genus Brucella (13). The omp2 locus contains two gene copies (omp2a and omp2b) coding for porin proteins that are used for molecular typing and identification of Brucella at the species, biovar, or strain level (14). The point mutations may effect the sequence amplified and change the diagnosis sites of specific restriction enzymes (15). The PCR-RFLP method has several advantages in comparison to bacteriological techniques. It is considered rapid and easy to performed, cost effective, easy to fit training into laboratory schedules, and does not need advanced tools and live specimens. Disadvantages include need for specific restriction enzymes and failure to identify variations in the nucleic acid sequence analyses (16). This study was the first report to determine the molecular typing of Brucella species by PCR-RFLP in Hamadan, west of Iran. The aim of this study was to analyze the epidemiological correlation and molecular typing of Brucella spp isolated from human and animals using the PCR-RFLP method.
2. Objectives
This study aimed at identifying genetic polymorphisms of omp2a genes among 90 Brucella spp. isolated from human and animals using the PCR-RFLP method. Also, it evaluated the genetic diversity and relationship among Brucella strains using PCR – RFLP and gene sequencing.
3. Methods
3.1. Samples Collection
In total, 90 Brucella strains were isolated from different sources of humans and animals (cattle, sheep and goat) in two regions of Iran (Tehran and Hamadan). Sixty Brucella spp were isolated from blood cultures collected from patients, who were referred to Sina Hospital of Hamadan with clinical symptoms, including malaise, arthralgia, myalgia, weakness, weight loss, splenomegaly, lymphadenopathy, and serum antibody titers ≥ 1/160. Ten milliliters of blood from each patient was transferred to the BACTEC blood culture system (9050 BD Company, U.S.A) and incubated at 37°C for seven day (17). Thirty animal Brucella isolates were obtained from the Department of Microbiology of the Faculty of Medicine, Iran University of Medical Sciences, Tehran, Iran. These bacteria were isolated from blood, retropharyngeal, and lymph nodes of animals (cattle, sheep, and goat) that had seropositive tests for brucellosis.
3.2. Bacteriological Identification
Brucella strains were grown on 5% sheep Brucella‐agar (Merck, Germany) plates and incubated at 37°C in the presence of 5% to 10% CO2 for 72 hours. Morphology and biochemical reactions including catalase, oxidase, urease hydrolysis tests, CO2 requirement and H2S production was considered (7). All isolates were cultured on Brucella broth media (Merck, Germany) with 20% glycerol and stored at -80°C in the department of microbiology of Hamadan University of Medical Sciences, Hamadan, Iran.
3.3. Polymerase Chain Reaction Assay
DNA was extracted by a modification of the method described by Queipo-Ortuno (18). The Brucella isolates were examined for the presence of omp2a gene by PCR amplification (19). Polymerase chain reaction for omp2a was performed in the thermal cycler (Bio-Rad, USA). The Primer sequences and condition of PCR is shown in Tables 1 and 2.
Polymerase Chain Reaction Mixture
| Reaction | Volume, µL | Company |
|---|---|---|
| Mastermix 2X | 12.5 | Pars Tous Co, IRAN |
| DN A template | 2 | - |
| Primer forward 10 pmol | 1 | Bioneer Co, Korea |
| Primer reverse 10 pmol | 1 | Bioneer Co, Korea |
| DDH2O | 8.5 | - |
| Total volume | 25 | - |
Primers Sequences, Temperature Program, and Cycling Conditions for Omp2a –Polymerase Chain Reaction
The PCR products were analyzed using the electrophoresis technique on 1.5% agarose gel in 1x Tris-Boric-EDTA (TBE) buffer. Amplified products were stained by syber-safe and visualized under UV transilluminator. Brucella abortus S19 strain was used as a positive control and sterile distilled water was used as a negative control. A 1-kb plus ladder (Bioneer Co., Korea) was used as the molecular size marker (20).
3.4. Digestion of the Polymerase Chain Reaction Products
Polymerase chain reaction-RFLP was used to differentiate all strains of Brucella. The PCR products were digested using Pst I restriction endonuclease enzyme. The PstI restriction enzyme was used according to the manufacturer’s (Thermo, Fermentase, USA) instructions. Briefly, the digestion mixture included 5 μL of PCR product, 1 μL enzyme buffers, 0.5 μL PstI restriction enzyme (10 to 20 U), and sterile distilled water up to 15.5 μL. The microtubes were incubated for four hours at 37°C. The products of RFLP were electrophoresed on 1.5% agarose gel with Syber-safe stain, and gel visualized under UV transilluminator. A 100-bp plus DNA ladder (Bioneer Co., Korea) was used as the molecular size marker (10).
3.5. PCR-RFLP analysis
Analysis of RFLP patterns was conducted using BioNumerics version 7.5 and compared the Dice coefficient and unweighted pair group method using arithmetic averages (UPGMA) groups with 0.5% optimization and a 0.5-% band position tolerance window (21).
4. Results
Ninety Brucella isolates, including 60 (66.7%) human strains comprised of B. melitensis, and 30 (33.3%) animal strains, including 28 B. melitensis and 2 B. abortus, were obtained from different sources and investigated. Phenotypic and molecular typing methods were performed to evaluate the species and the biovar of the Brucella isolates. The morphology and biochemical tests of the isolates showed that they had small colonies with high transparent and smooth surfaces on Brucella agar and gram-negative coccobacilli with Gram stain, were negative for H2S production, and positive for urease and oxidase without any necessity for the presence of CO2 and were identified as B. melitensis. However, the isolates, which were identified as B. abortus, were positive for oxidase, H2S production, urea hydrolysis, and growth in the presence of CO2.
4.1. Polymerase Chain Reaction Results
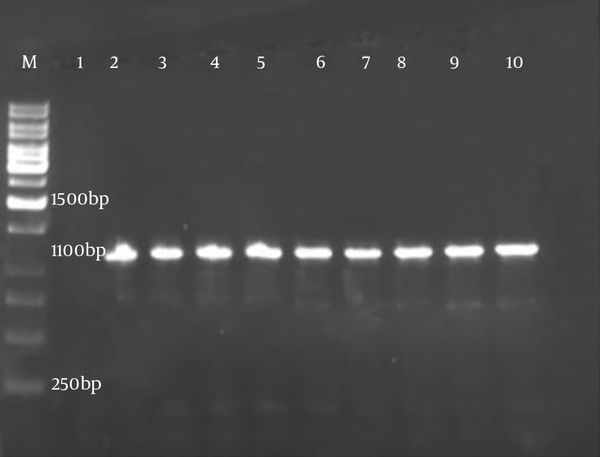
After identification of Brucella strains by biochemical standard methods, the isolates were confirmed by the PCR technique with obtained bands of 1100 bp for omp2a gene (Figure 1).
The result of omp2a gene amplification in human and animal isolates. Lane M: molecular ladder 1 kb, lane 1: negative control, lane 2 - 5: animal isolates of Brucella, lane 6 - 9: H human isolates of Brucella and lane 10: positive control (Brucellaabortus vaccine strain S19).
4.2. PCR-RFLP
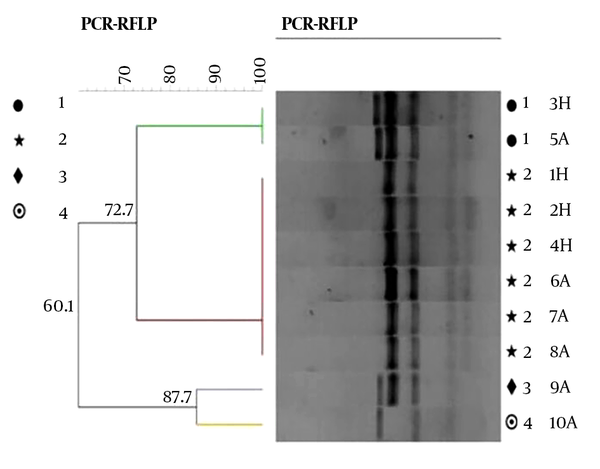
According to RFLP dendrogram, four groups (G1 to G4) and four patterns included P1 (five bands), P2 (four bands), P3 (four bands), and P4 (three bands), based on the host sources, were obtained (Figure 2). The created bands ranged from 60 to 500 bp. The fragments created in each pattern and the size of their bands is shown in Table 3. Pst1 digestion of the PCR products produced four distinct RFLP patterns for animal strains (P1 to P4) and two RFLP patterns (P1 to P2) for human strains (Figure 3).
Dendrogram of omp2a Gene PCR-RFLP. H: human isolates; A: animal isolates.
| RFLP Pattern Fragments, bp | P1 (60, 110, 270, 420, 460) | P2 (60, 110, 300, 450) | P3 (110, 280, 400, 500) | P4 (140, 290, 500) | Total | |
|---|---|---|---|---|---|---|
| B. melitensis strain 20236 | B. abortus | B. melitensis strain 20236 | B. melitensis Biovar1 | B. melitensis Biovar1 | ||
| Human isolates | 24 (40) | - | 36 (60) | - | - | 60 |
| Animals isolates | 8 (26.7) | 2 (6.6) | 11 (36.7) | 6 (20) | 3 (10) | 30 |
| Total | 34 (37.8) | 47 (52.2) | 6 (6.7) | 3 (3.3) | 90 | |
Agarose gel electrophoresis of pstI restriction fragments of omp2a polymerase chain reaction products. Lanes:1, 2, 4, 6, 7, 8 (fragments size: 60, 110, 300, 450 bp (P2)); lanes 3, 5 (fragments size: 60, 110, 270, 420, 460 bp (P1)); lane: 9 (fragments size: 110, 280, 400, 500 bp (P3)); lane: 10 (fragments size: 140, 290, 500 bp (P4)); lane M: 100 bp molecular weight marker; lanes: 1 - 3 human isolates and lanes: 4 - 10 animal isolates.
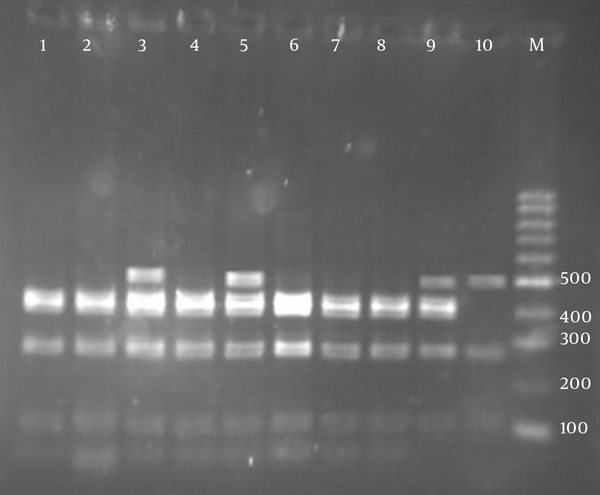
The omp2a gene sequence of isolates were submitted to GenBank and assigned by accession number: 06043563. The PCR-RFLP analysis of omp2a gene is displayed in Table 3.
By gene sequencing analysis of 60 human strains, 100% of isolates belonged to B. meltensis strain 20236 (P1, P2). Out of 30 animal Brucella isolates, 19 (63.4%) belonged to B. meltensis strain 20236 (P1, P2), nine (30%) belonged to B. meltensis biovare1 (P3, P4), and two (6.6 %) B. abortus (P1) (Table 3).
The dendrogram groups were defined by a cutoff value of 80% similarity, calculated using the BioNumerics software package (21).
From a total of 90 Brucella isolates, 24 (26.6%) of the human strains (B. melitensis), 10 (11%) of the animal strains (B. melitensis and B. abortus), and B. abortus vaccine strain (S19) were included in the P1 pattern (Table 3) with whole genetic similarity. Group 2 (P2) contained 36 (40%) of human (B. melitensis) and 11 (12.2%) of animal strains (B. melitensis) with complete genetic similarity. Group 3 (P3) and group 4 (P4) contained 6 (6.7%) and 3 (3.3%) of animal strains (B. melitensis), respectively. Group 2 (52.2%) had the most common patterns in all Brucella isolates. The dendrogram results showed a small variation among the isolates.
5. Discussion
Brucellosis is an important bacterial zoonosis, causing health problems in developing countries. Brucella are gram negative and facultative intracellular bacteria and are the causative agent of brucellosis that spread in various animal species and humans (22). The current research studied 60 human and 30 animal Brucella isolates. After identification of Brucella strains by phenotypic and biochemical tests, the isolates were confirmed for the presence of omp2a gene by PCR. All strains were omp2a gene positive. Recently, the PCR technique has increasingly been performed as a diagnostic and a confirmation method in Brucella spp. Identification (23, 24). In this study, detection of polymorphism for all Bruclla isolates by the omp2a gene as a target of PCR-RFLP and Pst1 enzyme was carried out.
For detection of polymorphism in Brucella spp. omp2a and omp2b fragments and several restriction enzymes were used in various studies (10, 25). In the present study, similar to other studies in Iran and different parts ofthe world, amplification of omp2a gene from clinical and animal isolates showed different sizes (50 to 500 bp) (1, 26). The study conducted by Unver et al. showed that the sizes of PCR products for clinical isolates of Brucella spp. ranged from 77 bp to 1200 bp (26).
In the present study, RFLP analysis of the omp2a gene in 90 Brucella isolates indicated four distinct RFLP patterns in animal isolates and two RFLP profiles in human isolates (Figure 3). The most common pattern in Brucella spp. isolated from human and animals in Hamadan and Tehran was pattern P2. Gene sequencing analysis for Brucella strains showed two profiles including, B. melitensis strain 20236 with P1 and P2 RFLP patterns in animal and human Brucella strains and B. melitensis biovar 1 with P3 and P4 RFLP patterns only in animal isolates. These results indicated that PCR-RFLP of omp2a gene was not able to accurately discriminate human and animal B. melitensis biovars from each other and from B. abortus. Also, comparable results were reported by Mirnejad et al. and Pishva et al. (6, 10).
In this study, using gene sequencing, B. melitensis strain 20236 was predominant yet in some other studies, B. melitensis biovar 1 was the predominant cause of human and animal brucellosis and it was commonly isolated from patients and widely spread in some areas of Iran (1, 6, 27). It has been reported that mixing herds and keeping the animals in shelters is a major risk factor for transmission of the infection (28). Iran’s geographical position has always been an important risk factor in the spread of brucellosis, mainly from eastern and western neighbors, such as Afghanistan, Pakistan, and Iraq, and it is possible for new Brucella strains to enter the country (29). These events may alter the epidemic genotypes of the disease in Iran and presence of new emerging strains and their source should be continuously considered.
In the current study, according to RFLP dendrogram, group 1 (P1) included B. melitensis isolated from humans and animals, and B. abortus strains isolated from animals. Group 2 (P2) contained B. melitensis isolated from humans and animals. Analysis of obtained patterns indicated low heterogeneity in the omp2 gene of Brucella strains, which may be due to common clone and same ancestor of Brucella spp. It is also possible that the bacteria had spread from animals to other animals or humans, and isolates were epidemiologically linked and therefore the outbreaks were likely due to the same species clone. Occurrence of these events can be directly due to the situation of brucellosis in animals in relation to the geographical area, and disease mostly involves people, who are in close contact with animals, such as veterinarian, ranchers and those, who are accustomed to eating goat, sheep, and raw milk (29). Therefore, defining the origin of infection in human and animals is also very important.
In this study, according to the results of sequencing, groups 3 (P3) and 4 (P4) isolated from animals belonged to B. melitensis biovare1, and another important result indicated complete similarities were not found among the animal isolates in single clones (G3 and G4). Furthermore, the B. abortus isolates did not transfer to human through animals, and this is similar to previous reports from Iran (30, 31). The current results showed different patterns of omp2a gene in Brucella spp. isolated from animals, and polymorphism analysis in omp2a gene will be useful in control and prevention of infections caused by Brucella strains and must be performed continuously.
5.1. Conclusions
The current findings, like other studies, confirmed that the frequency of B. melitensis is higher in human and animal resources and demonstrated that the B. melitensis strain 20236 was more predominant. Despite the broad application of PCR-RFLP in determining the polymorphism and understanding the epidemiology of important human pathogens, this technique was unable to differentiate human and animal species of B. melitensis from B. abortus, yet gene sequencing analysis could discriminate B. melitensis biovars.
Acknowledgements
References
-
1.
Salehi M, Pishva E, Salehi R, Rahmani R. Isolation of Brucella abortus using PCR-RFLP analysis. Iran J Public Health. 2006;35(4):22-7.
-
2.
Atluri VL, Xavier MN, de Jong MF, den Hartigh AB, Tsolis RM. Interactions of the human pathogenic Brucella species with their hosts. Annu Rev Microbiol. 2011;65:523-41. [PubMed ID: 21939378]. https://doi.org/10.1146/annurev-micro-090110-102905.
-
3.
Hasanjani Roushan MR, Mohrez M, Smailnejad Gangi SM, Soleimani Amiri MJ, Hajiahmadi M. Epidemiological features and clinical manifestations in 469 adult patients with brucellosis in Babol, Northern Iran. Epidemiol Infect. 2004;132(6):1109-14. [PubMed ID: 15635968]. [PubMed Central ID: PMC2870202].
-
4.
Zeinali M, Shirzadi M, Sharifian J. National guideline for Brucellosis control. Tehran: Ministry of Health and Medical Education; 2009. p. 10-7.
-
5.
Diaz Aparicio E. Epidemiology of brucellosis in domestic animals caused by Brucella melitensis, Brucella suis and Brucella abortus. Rev Sci Tech. 2013;32(1):43-51. 53-60. [PubMed ID: 23837364].
-
6.
Pishva E, Salehi R, Hoseini A, Kargar A, Taba FE, Hajiyan M, et al. Molecular typing of Brucella species isolates from Human and livestock bloods in Isfahan province. Adv Biomed Res. 2015;4:104. [PubMed ID: 26261806]. [PubMed Central ID: PMC4513321]. https://doi.org/10.4103/2277-9175.157798.
-
7.
Alton GG, Jones LM, Angus RD, Verger JM. Techniques for the brucellosis laboratory. 1988. Paris: Institut National de la Recherche Agronomique; 1988.
-
8.
Al Dahouk S, Tomaso H, Prenger-Berninghoff E, Splettstoesser WD, Scholz HC, Neubauer H. Identification of brucella species and biotypes using polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP). Crit Rev Microbiol. 2005;31(4):191-6. [PubMed ID: 16417200]. https://doi.org/10.1080/10408410500304041.
-
9.
Hajia M, Fallah F, Angoti G, Karimi A, Rahbar M, Gachkar L, et al. Comparison of Methods for Diagnosing Brucellosis. Lab Med. 2013;44(1):29-33. https://doi.org/10.1309/lm4j9mwobipa6rbn.
-
10.
Mirnejad R, Mohammadi M, Majdi A, Taghizoghi N, Piranfar V. Molecular Typing of Brucella melitensis and B. abortus from Human Blood Samples Using PCR-RFLP Method. Jundishapur J Microbiol. 2013;6(6). https://doi.org/10.5812/jjm.7197.
-
11.
Nagalingam M, Shome R, Balamurugan V, Shome BR, NarayanaRao K, Vivekananda, et al. Molecular typing of Brucella species isolates from livestock and human. Trop Anim Health Prod. 2012;44(1):5-9. [PubMed ID: 21647774]. https://doi.org/10.1007/s11250-011-9886-1.
-
12.
De Massis F, Ancora M, Atzeni M, Rolesu S, Bandino E, Danzetta ML, et al. MLVA as an Epidemiological Tool To Trace Back Brucella melitensis Biovar 1 Re-Emergence in Italy. Transbound Emerg Dis. 2015;62(5):463-9. [PubMed ID: 26194658]. https://doi.org/10.1111/tbed.12397.
-
13.
Garcia-Yoldi D, Marin CM, Lopez-Goni I. Restriction site polymorphisms in the genes encoding new members of group 3 outer membrane protein family of Brucella spp. FEMS Microbiol Lett. 2005;245(1):79-84. [PubMed ID: 15796983]. https://doi.org/10.1016/j.femsle.2005.02.026.
-
14.
Pishva E, Salehi M, Gharavi M. Relationship between Brucella immunocomplex and glomerulopathies. Arch Clin Infect Dis. 2008;3(3).
-
15.
Lukinmaa S, Nakari UM, Eklund M, Siitonen A. Application of molecular genetic methods in diagnostics and epidemiology of food-borne bacterial pathogens. APMIS. 2004;112(11-12):908-29. [PubMed ID: 15638843]. https://doi.org/10.1111/j.1600-0463.2004.apm11211-1213.x.
-
16.
Berg H. Restriction Fragment Length Polymorphism Analysis of PCR-Amplified Fragments (PCR-RFLP) and Gel Electrophoresis - Valuable Tool for Genotyping and Genetic Fingerprinting. InTechopen; 2012. https://doi.org/10.5772/37724.
-
17.
Hashemi SH, Alikhani MY, Naseri Z, Farajnia S, Peeri-Dogaheh H. Diagnosis of human brucellosis by blood culture (BACTEC) and PCR method via whole blood and serum. Jundishapur J Microbiol. 2013;6(3):248-51.
-
18.
Queipo-Ortuno MI, Morata P, Ocon P, Manchado P, Colmenero JD. Rapid diagnosis of human brucellosis by peripheral-blood PCR assay. J Clin Microbiol. 1997;35(11):2927-30. [PubMed ID: 9350761]. [PubMed Central ID: PMC230089].
-
19.
Bahmani N, Naseri Z, Alikhani MY, Karami M, Kalvandi R. Evaluation of antibacterial effects of Withania coagulans and Cynara cardunculus extracts on clinical isolates of Brucella strains. Int J Med Res Health Sci. 2016;5(9):90-5.
-
20.
Mullis KB, Faloona FA. Specific synthesis of DNA in vitro via a polymerase-catalyzed chain reaction. Methods Enzymol. 1987;155:335-50. [PubMed ID: 3431465].
-
21.
Hunter PR, Gaston MA. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J Clin Microbiol. 1988;26(11):2465-6. [PubMed ID: 3069867]. [PubMed Central ID: PMC266921].
-
22.
Cerekci A, Kilic S, Bayraktar M, Uyanik MH, Yasar E, Esen B. [Comparison of conventional methods and real-time multiplex polymerase chain reaction for identification and typing of Brucella isolates of human origin]. Mikrobiyol Bul. 2011;45(3):392-400. [PubMed ID: 21935772].
-
23.
Kaynak-Onurdag F, Okten S, Sen B. Screening Brucella spp. in bovine raw milk by real-time quantitative PCR and conventional methods in a pilot region of vaccination, Edirne, Turkey. J Dairy Sci. 2016;99(5):3351-7. [PubMed ID: 26971148]. https://doi.org/10.3168/jds.2015-10637.
-
24.
Qasem JA, AlMomin S, Al-Mouqati SA, Kumar V. Characterization and evaluation of an arbitrary primed Polymerase Chain Reaction (PCR) product for the specific detection of Brucella species. Saudi J Biol Sci. 2015;22(2):220-6. [PubMed ID: 25737656]. [PubMed Central ID: PMC4336441]. https://doi.org/10.1016/j.sjbs.2014.09.014.
-
25.
Samadi A, Ababneh MM, Giadinis ND, Lafi SQ. Ovine and Caprine Brucellosis (Brucella melitensis) in Aborted Animals in Jordanian Sheep and Goat Flocks. Vet Med Int. 2010;2010:458695. [PubMed ID: 21052561]. [PubMed Central ID: PMC2971571]. https://doi.org/10.4061/2010/458695.
-
26.
Unver A, Erdogan HM, Atabay HI, Sahin M, Celebi O. Isolation, identification, and molecular characterization of Brucella melitensis from aborted sheep fetuses in Kars, Turkey. Revue de medecine veterinaire. 2006;157(1):42.
-
27.
Behroozikhah AM, Bagheri Nejad R, Amiri K, Bahonar AR. Identification at biovar level of Brucella isolates causing abortion in small ruminants of iran. J Pathog. 2012;2012:357235. [PubMed ID: 23251805]. [PubMed Central ID: PMC3515911]. https://doi.org/10.1155/2012/357235.
-
28.
Aitken I. Diseases of sheep. John Wiley & Sons; 2008.
-
29.
Esmaeili H. Brucellosis in Islamic republic of Iran. J Med Bacteriol. 2015;3(3-4):47-57.
-
30.
Najafi N, Ghassemian R, Davoody AR, Tayebi A. An unusual complication of a common endemic disease: clinical and laboratory aspects of patients with brucella epididymoorchitis in the north of Iran. BMC Res Notes. 2011;4:286. [PubMed ID: 21834966]. [PubMed Central ID: PMC3170334]. https://doi.org/10.1186/1756-0500-4-286.
-
31.
Shareef JM. A Review of Serological Investigations of Brucellosis Among Farm Animals and Humans in Northern Provinces of Iraq (1974?2004). J Vet Med Series B. 2006;53(s1):38-40. https://doi.org/10.1111/j.1439-0450.2006.01021.x.
