Abstract
Background:
The number of extended-spectrum β-lactamase (ESBL)-producing Enterobacteriaceae reported cases all over the world has continued to increase faster than the other resistance mechanisms, particularly in E. coli and K. pneumoniae.Objectives:
This cross-sectional study was designed to assess the prevalence of multidrug resistance of ESBL-producing urine isolates of E. coli and K. pneumoniae collected in Tehran hospitals, as well as the molecular characterizations of some ESBL genes, with an emphasis on occurrence rates by sex.Materials and Methods:
A total of 190 E. coli and K. pneumoniae isolates were collected from patients’ urine samples in hospitals from Tehran, Iran during 2009 - 2010, and were screened for antibiotic susceptibility, ESBL phenotype, and presence of blaCTX-M and blaTEM genes. Minimal inhibitory concentration (MIC) for ceftazidime and cefotaxime were made by agar dilution method.Results:
The ESBL phenotype was detected in 55.5% of E. coli and 46.4% of K. pneumoniae isolates. Presence of blaCTX-M-1 was dominant in both organisms. The prevalence of blaCTX-M-1 carrying isolates among ESBL-producing K. pneumoniae and E. coli isolates were 49.1% and 85.7%, respectively. Among ESBL-producing isolates, 68.5% of E. coli and 59.3% of K. pneumoniae isolates carried the blaTEM genes, and simultaneous carrying of blaCTX-M and blaTEM genes was observed in 68.5% of E. coli and 33.3% of K. pneumoniae isolates. The resistant rate to ceftazidime, cefotaxime, and cefepime was significantly higher in K. pneumoniae and E. coli isolates from male patients urine samples. A significant higher rate of blaCTX-M-1, blaTEM, and co-blaCTX-M-1-blaTEM genes were seen for E. coli and K. pneumoniae isolates in male patients’ urine.Conclusions:
The results indicate that the rates of ESBLs are high in urine E. coli and K. pneumoniae isolates from Tehran hospitals. Also this study indicates that the urine isolates from male patients are significantly more resistant than the female isolates.Keywords
1. Background
Urinary tract infection (UTI) is the most common bacterial infections worldwide, and a frequent finding in general clinical practice (1, 2). This infection was diagnosed originally by the presence of at least 105 colony-forming units (CFU) of a single uropathogen in a urine specimen. However, in recent years, the cut-off limit has been reduced as bacterial count of ≥ 103 and 102 CFU/mL (2, 3). UTI accounted for 25% - 40% of the nosocomial infections, and approximately 80% of cases associated with the use of urinary catheters (4, 5).
Escherichia coli and Klebsiella pneumoniae are the most important causal agents of Gram-negative bacteriuria both in hospital and community acquired UTIs (2, 6). Extended-spectrum β-lactamase (ESBL)-producing E. coli and K. pneumoniae are an emerging cause of UTI worldwide, often resistant to commonly prescribed antimicrobial agents (7, 8). The prevalence of ESBLs in clinical E. coli and K. pneumoniae isolates in Iran has been found to be 21% - 56% and 12% - 69.7%, respectively (9-15). TEM and CTX-M are most common kinds of ESBLs worldwide. Phylogenetically, CTX-M enzymes have been classified into 5 major groups based on their amino acid similarities: the CTX-M-1 cluster (CTX-M-1, -3, -10, -11, -12, -15, -28, and FEC-1), the CTX-M-2 cluster (CTX-M-2, -4, -5, -6, -7, -20, and TOHO-1), the CTX-M-8 cluster (CTX-M-8), the CTX-M-25 cluster (CTXM- 25 and -26), and the CTX-M-9 cluster (CTX-M-9, -13, -14,-16, -17, -19, -21, -24, -27, and TOHO-2). The extraordinary dissemination of the blaCTX-M genes in mobile genetic elements, including plasmids and transposons worldwide has been referred as the CTX-M pandemic (16). The co-resistance occurrence, particularly to aminoglycosides and fluoroquinolones was observed in CTX-M producing organisms (17, 18). The TEM-type ESBLs are derivatives of TEM-1 and TEM-2. They can hydrolyze third-generation cephalosporins and are inhibited by clavulanic acid (19). This study was conducted to assess the prevalence of multidrug resistance of ESBL-producing clinical isolates of E. coli and K. pneumoniae in Tehran hospitals as well as the molecular characterizations of some ESBL genes, with an emphasis on occurrence rates by sex.
2. Materials and Methods
2.1. Clinical Isolates
A total of 127 K. pneumoniae and 63 E. coli isolates were collected from different teaching hospitals in Tehran, Iran, during 2009 - 2010. These isolates were taken from male (n = 62) and female urine cultures (n = 128). The isolates were identified by conventional biochemical tests as K. pneumoniae and E. coli.
2.2. Susceptibility Tests and Confirmation of ESBL Production
Susceptibility testing was conducted by disk diffusion according to the guidelines of the clinical and laboratory standards institute (CLSI). Twelve antimicrobial disks (Mast group; UK) included amoxicillin, cefoxitin, ceftazidime, cefotaxime, cefepime, aztreonam, gentamicin, amikacin, tetracycline, co-trimoxazole, imipenem, and ciprofloxacin. E. coli ATCC 25922 was used for quality control purposes in susceptibility testing. The ESBL phenotype was detected by combined disk methods using disks of ceftazidime and cefotaxime with and without clavulanic acid. The MICs was determined by agar dilution method using ceftazidime and cefotaxime with (4 mg/L) and without clavulanic acid (20).
2.3. Polymerase Chain Reaction Amplification of ESBL Genes
The specific primers for diverse CTX-M groups (CTX-M-1, CTX-M-2, and CTX-M-9 groups) and TEM β-lactamase, as described previously, were used (21, 22). These specific primer pairs were as follows: M1-F:5’-GGTTAAAAAAT CACTGCGTC-3’ and M1-R: 5’-TTGGTGACGATTTTAGCCGC-3’ (for blaCTX-M-1, amplicon size: 864 bp) , M9-F: 5’-ATGGTGACAAAGAGAGTGCA-3’ and M9-R: 5’-CCCTTCGGCGATGATTCTC-3’ (for blaCTX-M-9, amplicon size: 869), M2-F: R: 5’- ATGATGACTCAGAGCATTCG -3’, M2-R: 5’- CCCTTCGGCGATGAT TCTC -3’ (for blaCTX-M-9, amplicon size: 869), and TEM-F: 5’- ATGAGTATTCAA CATTTCCG -3’, TEM-R: 5’- CCAATGCTTAATCAGTGAGG -3’ (for blaTEM, amplicon size: 850). Amplification reactions were performed in a total volume of 25 µL of reaction mixture containing 5 µL of 10 × PCR buffer, 2.5 mM Mgcl2, 200 µM dNTPs, 1.25 units of Taq polymerase, 10 pmol of each primer, and 1 µL of sample DNA. Amplification reactions were carried out in an Eppendorf thermal cycler (Eppendorf AG, Hamburg, Germany), with an initial denaturation (4 minutes at 94°C), followed by 30 cycles of denaturation (60 seconds at 94°C), annealing (30 seconds at 55°C), and extension (1 minute at 72°C), with a single final extension of 5 minutes at 72°C. DNA template from control clinical strains with well characterized CTX-M (groups 1, 2, 9) and TEM β-lactamases were used as the positive controls for PCR (23). The PCR products were analyzed on 1% agarose gels stained with ethidium bromide.
2.4. Statistical Analysis
Comparisons of proportions were tested using the χ2-test with SPSS version 19. A value of P < 0.05 was considered significant.
3. Results
3.1. Antimicrobial Susceptibility
The antibiogram results are shown in Table 1. As shown, maximal resistance in both microorganisms was found against amoxicillin (89.7% and 92%), followed by ceftazidime (51.1% and 55.5%), cefotaxime (50.3% and 71.4%), and cefoxitin (61.4% and 41.2%). Only 8.6% of K. pneumoniae isolates were resistant to ciprofloxacin, while with E. coli isolates, the rate of ciprofloxacin resistance was 36.5% (Table 1). Although the resistance rates to tetracycline (79.3%) and co-trimoxazole (69.1%) were high in E. coli isolates, these were almost moderate in K. pneumoniae isolates (36.2% and 30.1%, respectively). The resistance rate to other antibiotics was almost similar and imipenem was effective against for all K. pneumoniae and E. coli isolates. Resistance rate differences could be related to the sex, although it was not significant in all cases. According to the Table 1, drug resistance in both bacteria isolates from male patients’ urine was higher than the female isolates. The resistance rate to ceftazidime, cefotaxime, cefepime, aztreonam, and gentamicin was significantly (P < 0.05) higher in K. pneumoniae isolates from male patients’ urine. In E. coli isolates, significant (P < 0.05) differences were found to ceftazidime, cefotaxime, cefepime and co-trimoxazole. Co-resistance to all tested antibiotics (except imipenem) was detected in 6.3% (4 out of 63) of E. coli isolates, while, this multi-resistance pattern rates was 18.7% (3 out of 16) in male and 2.1% (1 out of 47) in female samples. In K. pneumoniae, simultaneous resistance to all tested drug (except imipenem) was detected only in 1 isolate from male and 1 from female samples.
Antimicrobial Resistance Rates of K. pneumoniae and E. coli isolates (Number and Percentage Resistant) by Disk Diffusion Methoda
| K. pneumoniae | E. coli | |||||
|---|---|---|---|---|---|---|
| Female (n = 81) | Male (n = 46) | Total (n = 127) | Female (n = 47) | Male (n = 16) | Total (n = 63) | |
| Amoxicillin | 71 (87.6) | 43 (93.4) | 114 (89.7) | 42 (89.3) | 16 (100) | 58 (92) |
| Aztreonam | 22 (27.1) | 24 (52.8) | 46 (36.2) | 14 (29.7) | 9 (56.2) | 23 (36.5) |
| Amikacin | 20 (24.6) | 14 (30.4) | 34 (26.7) | 9 (19.1) | 6 (37.5) | 15 (23.8) |
| Cefepime | 21 (25.9) | 23 (50) | 44 (34.6) | 11 (23.4) | 11 (68.7) | 22 (34.9) |
| Ceftazidime | 35 (43.2) | 30 (65.2) | 65 (51.1) | 22 (46.8) | 13 (81.2) | 35 (55.5) |
| Cefotaxime | 34 (41.9) | 30 (65.2) | 64 (50.3) | 29 (61.7) | 16 (100) | 45 (71.4) |
| Cefoxitin | 46 (56.7) | 32 (69.5) | 78 (61.4) | 17 (36.1) | 9 (56.2) | 26 (41.2) |
| Tetracycline | 28 (34.5) | 18 (39.1) | 46 (36.2) | 36 (76.5) | 14 (87.5) | 50 (79.3) |
| Ciprofloxacin | 5 (6.17) | 6 (13) | 11 (8.6) | 15 (31.9) | 8 (50) | 23 (36.5) |
| Gentamicin | 17 (20.9) | 22 (47.8) | 39 (30.7) | 11 (23.4) | 7 (43.7) | 18 (28.5) |
| Co-trimoxazole | 28 (34.5) | 22 (47.8) | 50 (39.3) | 25 (53.1) | 14 (87.5) | 39 (69.1) |
| Imipenem | 0 | 0 | 0 | 0 | 0 | 0 |
Of 127 K. pneumoniae isolates, 65 (51.1%) were resistant to ceftazidime (MIC > 4 mg/L) and 64 (50.3%) were resistant to cefotaxime (MIC > 1 mg/L). As shown in Table 2, the MIC90 was the lowest for cefotaxime (128 mg/L) compared with 265 mg/L for ceftazidime in K. pneumoniae isolates. About 55.5% and 71.4% of E. coli isolates were non-susceptible to ceftazidime and cefotaxime, respectively. The MIC90 for cefotaxime was the highest (512 mg/L) compared with 64 mg/L for ceftazidime in E. coli isolates (Table 2).
Minimum Inhibitory Concentrations of CAZ and CTX for K. pneumoniae and E. coli isolates
| MIC, mg/L | ||||
|---|---|---|---|---|
| Minimum | 50% | 90% | Maximum | |
| E. coli (n = 63) | ||||
| CTX | > 0.5 | 8 | 512 | < 512 |
| CAZ | > 0.5 | 8 | 64 | < 512 |
| K. pneumoniae (n = 127) | ||||
| CTX | > 0.5 | 1 | 256 | < 512 |
| CAZ | > 0.125 | 4 | 128 | < 512 |
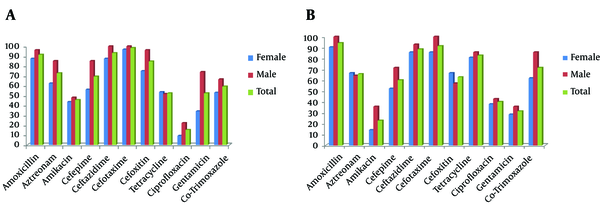
All resistant isolates to ceftazidime and cefotaxime were screened by combined disk test for ESBL production. The differences in inhibition zones of β-lactam disks with and without clavulanic acid were 8 to 23 mm. As shown in Figure 1, the ESBL phenotype was detected in 55.5% of E. coli and 46.4% of K. pneumoniae isolates. Co-resistances to amikacin, gentamicin, tetracycline, and co-trimoxazole were common in both ESBLs organisms. The relative resistance rate of the ESBL-producing K. pneumoniae to various antibiotics was higher than E. coli isolates, although some trends were observed: E. coli isolates were more resistant to ciprofloxacin, tetracycline, and co-trimoxazole. In ESBL producer, the MIC90 of cefotaxime and ceftazidime were 512 mg/L for E. coli and 256 mg/L for K. pneumoniae isolates. There was also a high rate of ESBL-producing E. coli (87.5% vs. 44.6%) and K. pneumoniae (58.6% vs. 39.5%) isolates from male patients’ urine. The difference of ESBL production in relation to sex was significant (P < 0.05) in E. coli isolates.
Antimicrobial Resistance Rate of ESBL-Producing Isolates (Percentage Resistant) by Disk Diffusion Method
3.2. Detection of Resistant Genes
Polymerase chain reaction (PCR) was performed for detection of CTX-M group genes in all cefotaxime resistant isolates according to the results obtained by antibiogram tests. Twenty-nine (46%) K. pneumoniae and 13 (66.6%) E. coli isolates carried the blaCTX-M-1 group alleles. The prevalence of blaCTX-M1 carrying isolates among ESBL-producing K. pneumoniae and E. coli isolates was 49.1% and 85.7%, respectively. Co-resistances to all tested β-lactams (except imipenem) among blaCTX-M-1 carrying K. pneumoniae and E. coli isolates was found in 21 (72.4%) and 10 (33.3%) isolates, respectively, and simultaneous resistance to other antibiotic families were more common. None of our isolates were positive for CTX-M groups 2 and 9.
The MICs for blaCTX-M1 carrying isolates were determined for ceftazidime and cefotaxime with and without inhibitor (Table 3). The MIC of ceftazidime and cefotaxime were 8 to 512 mg/L. The 58.6% of K. pneumoniae and 46.6% of E. coli isolates had high level resistance (≥ 256 mg/L) for cefotaxime, while for ceftazidime; this was 34.4% and 23.3%, respectively.
Minimum Inhibitory Concentrations of CAZ and CTX With and Without Clavulanate for blaCTX-M-1 Carrying Isolates
| mg/L | < 4 | 8 | 16 | 32 | 64 | 128 | 256 | 512 ≤ |
|---|---|---|---|---|---|---|---|---|
| E. coli (n = 30) | ||||||||
| CTX | 0 | 1 | 3 | 5 | 4 | 3 | 5 | 9 |
| CTX-CA | 30 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| CAZ | 4 | 3 | 9 | 4 | 3 | 0 | 1 | 6 |
| CAZ-CA | 21 | 1 | 1 | 2 | 1 | 0 | 0 | 0 |
| K. pneumonia (n = 29) | ||||||||
| CTX | 0 | 2 | 0 | 0 | 3 | 7 | 10 | 7 |
| CTX-CA | 20 | 5 | 1 | 3 | 0 | 0 | 0 | 0 |
| CAZ | 0 | 2 | 2 | 6 | 6 | 3 | 6 | 4 |
| CAZ-CA | 12 | 10 | 2 | 4 | 1 | 0 | 0 | 0 |
The phenotypic ESBL agar dilution confirmatory test for blaCTX-M-1 carrying isolates was positive, and the prevalence of ceftazidime and cefotaxime resistant isolates dramatically decreased from up to 3-6 dilution in the presence of clavulanic acid (Table 3).
All ESBL producer isolates were screened for the presence of blaTEM genes, in which 24 (68.5%) of E. coli and 35(59.3%) of K. pneumoniae isolates carried the blaTEM genes (Figure 2). Simultaneous carrying of blaCTX-M and blaTEM genes was observed in 24(68.5%) of E. coli and 20(33.3%) of K. pneumoniae isolates.
Prevalence of ESBLs and Resistance Genes in A, K. pneumoniae and B, E. coli Isolates According to the Patients’ Sex
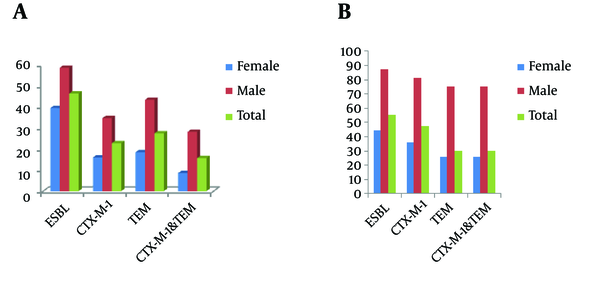
The presence of ESBL phenotype and resistant genes in both microorganisms according to the patient sex was shown in Figure 2. In E. coli and K. pneumoniae isolated from male patients’ urine samples, the resistant genes were found at least two fold higher than females’ isolates.
4. Discussion
The number of ESBL-producing Enterobacteriaceae reported cases all over the world has continued to rise faster than the other resistance mechanisms, particularly in E. coli and K. pneumoniae (1). These enzymes hydrolyze third-generation cephalosporins and are inhibited by clavulanic acid. Regarding the prevalence of the ESBLs genotype (e.g. CTX-M, TEM) and also geographical variation in the occurrence of different ESBL variants (e.g. CTX-M-1, CTX-M-9), the present study provides further data concerning urinary isolates of E. coli and K. pneumoniae (among them the relationship between the rate of resistance and patient sex) in Tehran, Iran.
The high ESBL occurrence determined for E. coli (55.5%) and K. pneumoniae (46.4%) in this study is nearly identical to the values found in previous studies conducted in Tehran (11, 12, 23). Surveillance data reported various levels of ESBL-producing strains of K. pneumoniae and E. coli throughout the world. The ESBL rates in northern Europe, north America, and Australia is 5%-10%, in contrast, high level ESBL producer K. pneumoniae and E. coli were found in Syria (≥ 60%), India (≥ 80%), China (≥ 60%), and other areas in east and southeast Asia, southern Europe (≥ 30%) (21, 22, 24).
The ESBL producers are frequently resistant to non-β-lactam antibiotics, including aminoglycosides and quinolones. In our ESBL isolates, high rates of resistance were found against amikacin, gentamicin, tetracycline, and co-trimoxazole, but the ciprofloxacin resistance was higher among E. coli (40%) compared to K. pneumoniae (15%) isolates. The ciprofloxacin resistance is mainly encoded chromosomally, while other co-resistances are often encoded by the same plasmids that determine the ESBL (8, 17, 19, 21).
The dramatic shifts reported in the types of ESBLs from Europe, Asia, and South America, with strains producing CTX-M becoming dominant (19, 21, 24, 25). The urinary ESBL E. coli and K. pneumoniae isolates in this study were carried high rate of blaCTX-M gene (46.7% and 22.8%, respectively) and blaTEM genes (30% and 27.5%, respectively). In K. pneumoniae isolates, the rate of blaTEM genes was slightly higher than blaCTX-M gene, in contrast, the blaCTX-M gene was predominant in E. coli, and all blaTEM carrying isolates, simultaneously contained blaCTX-M genes.
Regarding the geographical occurrence of specific CTX-M genotypes, blaCTX-M-1 is the only genotype that was detected in this study. In previous studies conducted in Tehran, the CTX-M-1 and CTX-M-3 were reported the prevalent CTX-M enzymes (10, 12, 23).
Regarding sex differences in the rate of resistance, higher rates of ESBL phenotype presence, blaCTX-M-1, blaTEM, blaCTX-M-1, and blaTEM gene were seen for E. coli and K. pneumoniae strains in male patients’ urine. Also, higher rates of resistance to amoxicillin, cefoxitin, ceftazidime, cefotaxime, cefepime, aztreonam, gentamicin, amikacin, tetracycline, co-trimoxazole, and ciprofloxacin were seen for both organisms in male patients’ urine. The exact reason for these differences is not clear; however, the faecal colonisation of resistance isolates in male patients could be responsible. Although, there is no studies to address this speculation.
The high resistance observed in this study could be due to the lack of a strict policy of antibiotic use in our country. The use of third and fourth generation cephalosporins has been reported the most important selective pressure in the appearance of different genotype and ESBL variants (25). The specific monitoring studies are needed to detect the association between specific antibiotic consumption and antimicrobial resistance.
In summary, CTX-M1 was dominant in E. coli and K. pneumoniae isolates and imipenem has remained quite active against more resistant strains. In addition, co-production of different ESBLs was frequently detected in both organisms. Finally, a high prevalence of ESBL genes was detected in E. coli and K. pneumoniae isolated from male patients’ urine.
Acknowledgements
References
-
1.
Saadeh SA, Mattoo TK. Managing urinary tract infections. Pediatr Nephrol. 2011;26(11):1967-76. https://doi.org/10.1007/s00467-011-1801-5.
-
2.
Schmiemann G, Gagyor I, Hummers-Pradier E, Bleidorn J. Resistance profiles of urinary tract infections in general practice--an observational study. BMC Urol. 2012;12:33. [PubMed ID: 23171154]. https://doi.org/10.1186/1471-2490-12-33.
-
3.
Giesen LG, Cousins G, Dimitrov BD, van de Laar FA, Fahey T. Predicting acute uncomplicated urinary tract infection in women: a systematic review of the diagnostic accuracy of symptoms and signs. BMC Fam Pract. 2010;11:78. [PubMed ID: 20969801]. https://doi.org/10.1186/1471-2296-11-78.
-
4.
Bagshaw SM, Laupland KB. The epidemiology of the intensive care unit acquired urinary tract infections. Curr Opin Infect Dis. 2006;19(1):67.
-
5.
Conway LJ, Larson EL. Guidelines to prevent catheter-associated urinary tract infection: 1980 to 2010. Heart Lung. 2012;41(3):271-83. [PubMed ID: 21925731]. https://doi.org/10.1016/j.hrtlng.2011.08.001.
-
6.
Wu YH, Chen PL, Hung YP, Ko WC. Risk factors and clinical impact of levofloxacin or cefazolin nonsusceptibility or ESBL production among uropathogens in adults with community-onset urinary tract infections. J Microbiol Immunol Infect. 2014;47(3):197-203. [PubMed ID: 23063776]. https://doi.org/10.1016/j.jmii.2012.09.001.
-
7.
Weisenberg SA, Mediavilla JR, Chen L, Alexander EL, Rhee KY, Kreiswirth BN, et al. Extended spectrum beta-lactamase-producing Enterobacteriaceae in international travelers and non-travelers in New York City. PLoS One. 2012;7(9). ee45141. [PubMed ID: 23028808]. https://doi.org/10.1371/journal.pone.0045141.
-
8.
Ko KS, Yeom JS, Lee MY, Peck KR, Song JH. Clonal dissemination of extended-spectrum beta-lactamase (ESBL)-producing Klebsiella pneumoniae isolates in a Korean hospital. J Korean Med Sci. 2008;23(1):53-60. [PubMed ID: 18303199]. https://doi.org/10.3346/jkms.2008.23.1.53.
-
9.
Behrooozi A, Rahbar M, Jalil V. Frequency of extended spectrum beta-lactamase (ESBLs) producing Escherichia coli and Klebseilla pneumonia isolated from urine in an Iranian 1000-bed tertiary care hospital. Afr J Microbiol Res. 2010;4(9):881-4.
-
10.
Nematzadeh S, Shahcheraghi F, Feizabadi MM, Nikbin VS, Nasehi L. Molecular characterization of CTX-Mbeta-lactamases among Klebsiella pneumoniae isolated from patients at Tehran hospitals. Indian J Med Microbiol. 2011;29(3):254-7. [PubMed ID: 21860105]. https://doi.org/10.4103/0255-0857.83908.
-
11.
Eftekhar F, Rastegar M, Golalipoor M, Mansoursamaei N. Detection of Extended Spectrum B-Lactamases in Urinary Isolates of Klebsiella pneumoniae in Relation to Bla, Bla and Bla Gene Carriage. Iran J Public Health. 2012;41(3):127-32. [PubMed ID: 23113157].
-
12.
Feizabadi MM, Delfani S, Raji N, Majnooni A, Aligholi M, Shahcheraghi F, et al. Distribution of bla(TEM), bla(SHV), bla(CTX-M) genes among clinical isolates of Klebsiella pneumoniae at Labbafinejad Hospital, Tehran, Iran. Microb Drug Resist. 2010;16(1):49-53. [PubMed ID: 19961397]. https://doi.org/10.1089/mdr.2009.0096.
-
13.
Derakhshan S, Najar Peerayeh S, Fallah F, Bakhshi B, Rahbar M, Ashrafi A. Detection of Class 1, 2, and 3 Integrons Among Klebsiella pneumoniae Isolated from Children in Tehran Hospitals. Arch Pediatr Infect Dis. 2013;1(4):164-8. https://doi.org/10.5812/pedinfect.11845.
-
14.
Memariani M, Najar Peerayeh S, Zahraei Salehi T, Shokouhi Mostafavi SK. Occurrence of SHV, TEM and CTX-M beta-Lactamase Genes Among Enteropathogenic Escherichia coli Strains Isolated From Children With Diarrhea. Jundishapur J Microbiol. 2015;8(4). ee15620. [PubMed ID: 26034531]. https://doi.org/10.5812/jjm.8(4)2015.15620.
-
15.
Najar Peerayeh S, Eslami M, Memaryani M, Siadat SD. High Prevalence of bla CTX-M-1 Group Extended-Spectrum β-lactamase Genes in Escherichia coli Isolates From Tehran. Jundishapur J Microbiol. 2013;6(7). https://doi.org/10.5812/jjm.6863.
-
16.
Canton R, Coque TM. The CTX-M beta-lactamase pandemic. Curr Opin Microbiol. 2006;9(5):466-75. [PubMed ID: 16942899]. https://doi.org/10.1016/j.mib.2006.08.011.
-
17.
Canton R, Ruiz-Garbajosa P. Co-resistance: an opportunity for the bacteria and resistance genes. Curr Opin Pharmacol. 2011;11(5):477-85. [PubMed ID: 21840259]. https://doi.org/10.1016/j.coph.2011.07.007.
-
18.
Peerayeh SN, Rostami E, Siadat SD, Derakhshan S. High rate of aminoglycoside resistance in CTX-M-15 producing Klebsiella pneumoniae isolates in Tehran, Iran. Lab Med. 2014;45(3):231-7. [PubMed ID: 25051075]. https://doi.org/10.1309/LMDQQW246NYAHHAD.
-
19.
Paterson DL, Bonomo RA. Extended-spectrum beta-lactamases: a clinical update. Clin Microbiol Rev. 2005;18(4):657-86. [PubMed ID: 16223952]. https://doi.org/10.1128/CMR.18.4.657-686.2005.
-
20.
Performance standards for antimicrobial susceptibility testing. Twenty-two informational supplement. Wayne: CLSI; 2012.
-
21.
Eckert C, Gautier V, Saladin-Allard M, Hidri N, Verdet C, Ould-Hocine Z. Dissemination of CTX-M-type β-lactamase among clinical isolates of Enterobacteriaceae in Paris, France. Antimicrob Agents Chemother. 2004;48(4):1249-55.
-
22.
Perez F, Endimiani A, Hujer KM, Bonomo RA. The continuing challenge of ESBLs. Curr Opin Pharmacol. 2007;7(5):459-69. [PubMed ID: 17875405]. https://doi.org/10.1016/j.coph.2007.08.003.
-
23.
Mirzaee M, Owlia P, Mansouri S. Distribution of CTX-M β-lactamase genes among Escherichia coli strains isolated from patients in Iran. Lab Med. 2009;40(12):724-7.
-
24.
AL-Subol I, Nihad Y. Prevalence of CTX-M, TEM and SHV Beta-lactamases in Clinical Isolates of Escherichia Coli and Klebsiella Pneumoniae Isolated From Aleppo University Hospitals, Aleppo, Syria. Arch Clin Infect Dis. 2015;10(2).
-
25.
Livermore DM. Current epidemiology and growing resistance of gram-negative pathogens. Korean J Intern Med. 2012;27(2):128-42. [PubMed ID: 22707882]. https://doi.org/10.3904/kjim.2012.27.2.128.