Abstract
Background:
Enterococcus spp. are part of the normal flora of humans and animals. The nosocomial pathogenicity of Enterococcus spp. has emerged in recent years and has caused great concern due to developing of resistance to many antimicrobial agents.Objectives:
The current study aimed to determine the resistance pattern and the type of virulence genes in Enterococcus spp. isolated from Milad hospital of Tehran, Iran.Materials and Methods:
The current observational study was conducted from Apr 2014 to Feb 2015 on a total of 149 Enterococcus species isolated from Milad hospital in Tehran, Iran. The antibiotic susceptibility pattern of the bacteria was determined by the disc diffusion method for eight antibiotics. Minimum inhibitory concentration (MIC) of vancomycin was also done using agar-dilution assay by clinical and laboratory standards institute (CLSI) recommendations. The sodA, esp, cyl, ace and gelE genes were detected by polymerase chain reaction (PCR) assay.Results:
About 37.5%, 73%, 86.6%, 35.8%, 69%, 60.8%, 45% and 79% of the isolates were resistant to vancomycin, tetracycline, gentamicin, chloramphenicol, ciprofloxacin, penicillin, ampicillin and erythromycin, respectively. MIC on 38% of the isolates was ≥ 256 µg/mL. Although, the prevalence of vancomycin-resistant Enterococcus (VRE) strains belonged to two species, E. faecium showed high resistance to a broad range of antibiotics. In total, 94 isolates were positive for esp, and 59, 48 and 3 isolates were positive for ace, cylA and gelE, Respectively.Conclusions:
The results of the current study designate the important role of medical samples as reservoirs of resistance inducing elements. Early detection of VRE with their virulence trait will help to prevent the spread of vancomycin resistant Enterococcus species. Supervision for antibiotic usage in hospitals, especially for last option antibiotics, can prevent the spread of resistant isolates and losing all treatment options in the future.Keywords
1. Background
Enterococcus spp. are part of the normal flora of gastrointestinal tract, biliary tract, anterior urethra and female genital tract in both human and animal. These are important global causes of nosocomial infection disease (1). Two sources of infections with Enterococcus spp. are proposed: first, infections may be caused by Enterococcus species of patient's flora; second, infections may be caused by Enterococcus spp. acquired from hospital environments. Two common Enterococcus species isolated from nosocomial infections are E. faecium and E. faecalis.Enterococcus strains resistant to different antibiotics are a great public health problem, especially species isolated from hospital-acquired infections (1, 2). Enterococcus faecalis is responsible for 80% - 90% and Enterococcus faecium for the remaining human enterococcal infections (3-5). In 2005, there were 7066 infectious cases caused by Enterococcus species in the UK, which 28% were antibiotic resistant (6, 7). These microorganisms are reported as the second leading cause of nosocomial urinary tract infections, and the third leading cause of nosocomial bacteremia in hospitalized patients (1, 2, 6). Vancomycin-resistant Enterococcus (VRE) is established as a significant nosocomial pathogen since it was first reported 20 years ago. VRE are one of the most important nosocomial pathogens worldwide; they are also one of the most troublesome pathogens in hospitals in the United States and are increasing in European health institutions (6, 8, 9). In addition, the prevalence of VRE among clinical Enterococcus isolates is increasing rapidly (9). Colonization with VRE can lead to serious diseases such as urinary tract infections, bacteremia and endocarditis which many of them such as VRE sepsis can be fatal (9-11). VRE have prolonged survival on hands, gloves, and environmental surfaces which consequently can be transmitted to the patients (12). It is believed that nosocomial enterococci might have virulence elements that increase their ability to colonize hospitalized patients (13, 14). Enterococcus spp. may carry various genes that directly or indirectly contribute to virulence (15, 16).
Virulence factors of Enterococcus spp. may contribute to competition with other bacteria, colonization of the host, resistance against defense mechanisms of the host and production of pathological changes directly through production of toxins or indirectly through induction of inflammation (17, 18).
The presence of genes encoding virulence factors including collagen-binding protein (ace), aggregation substance (asa1), cytolysin (cylA), endocarditis antigen (efaA), enterococcal surface protein (esp), gelatinase (gelE) and hyaluronidase (hyl), is analyzed in recent years (19, 20).
The extracellular surface protein (Esp), encoded by the esp gene, is a cell wall-associated protein which serves as adhesion for the pathogen to host tissue colonization and persistence in urinary tract infections (21),while aggregation substances encoded by asa1 are responsible for increased bacterial adhesion to renal tubular cells and heart endocardial cells (16).
2. Objectives
The current study aimed to determine the antimicrobial resistance pattern of Enterococcus spp. by disc diffusion test (DDT) and minimum inhibitory concentration (MIC), and investigate the prevalence of virulence genes encoding gelatinase (gelE), collagen adhesion (ace), cytolysin (cylA) and enterococcal surface protein (esp) in Enterococcus species isolated from Milad hospital in Tehran, Iran.
3. Patients and Methods
3.1. Sampling Method
In the current study, a total of 149 Enterococcus spp. isolates were collected from different wards, such as intensive care unit (ICU), emergency and surgery rooms, of Milad hospital in Tehran, from Apr 2014 to Feb 2015. Various clinical samples via urine, blood, pus, stool, fluids and aspirates were collected aseptically from admitted patients; then the samples were kept at 4°C until processing. It is noteworthy that, Milad hospital is the largest state specialized and sub-specialized hospital in Iran with 1000 licensed beds.
3.2. Isolation and Phonotypical Identification
The isolates were identified to genus and species level using clinical and laboratory standards institute (CLSI) guidelines. Specimens were transferred to the hospital laboratory and cultured on selective M-Enterococcus agar (Merck, Germany) with 2, 4 and 8 µg/mL vancomycin. All of the samples were cultured on bile-esculin-agar (Merck, Germany). Enterococcus genus was identified using colony morphology, Gram staining, catalase, pyrolidonyl aminopeptidase (PYR) and 6.5% salt tolerance tests. Species were identified using mannitol, sorbitol, arabinose, lactose, methyl-alpha-glucopyranoside and motility tests. Reactions were observed after 24 hours of incubation at 37°C. Results were later confirmed by PCR.
3.3. Antibiotic Susceptibility Tests
The antimicrobial susceptibility of the strains was determined using the disk diffusion method according to the CLSI guidelines for the following antimicrobial agents (Mast, UK): ampicillin (10 µg), penicillin (10 units), gentamicin (10 µg), ciprofloxacin (5 µg), erythromycin (15 µg), vancomycin (30 µg) tetracycline (30 µg) and chloramphenicol (30 µg) .Considering the increasing rate of vancomycin resistant Enterococcus species in clinical isolates worldwide, the MICs of vancomycin were determined using the agar dilution method based on the CLSI (2013) guidelines. Enterococcus faecalis (ATCC 29212) was used as control strain to perform antimicrobial tests. Vancomycin was tested in the range of 0.25 - 256 µg/mL. All of the tests were performed in duplicate. Muller Hinton agar was supplemented with different concentrations of vancomycin. Ten microliters of bacterial culture was spot inoculated after adjusting the turbidity with McFarland 0.5 standard. The plates were incubated at 37°C for 24 hours and examined for growth. MIC break points recommended by the CLSI guidelines were used. MIC ≤ 4 was considered as sensitive, MIC between 4 - 32 as intermediate resistant, MIC ≥ 32 as resistant and MIC ≥ 256 was considered as highly resistant to vancomycin.
3.4. Polymerase Chain Reaction
PCR assay based on the specific detection of sodA genes (Table 1) was used to confirm the identification of E. faecalis and E. faecium.
The Primer Sequences and Expected Size of Amplicons for Each PCR Result
| Primer Name | Target Gene | Oligonucleotide Sequences | Product Size, bp |
|---|---|---|---|
| sodA-fcm | sodA | F: TTGAGGCAGACCAGATTGACG; R: TATGACAGCGACTCCGATTCC | 658 |
| sodA-fcl | sodA | F: ATCAAGTACAGTTAGTCT; R: ACGATTCAAAGCTAACTG | 941 |
| gelE | gelE | F: TATGACAATGCTTTTTGGGAT; R: ATGACAATGCTTTTTGGGAT | 213 |
| esp | esp | F: AGATTTCATCTTTGATTCTTGG; R: AATTGATTCTTTAGCATCTGG | 510 |
| cylA | cylA | F: ACTCGGGGATTGATAGGC; R: GCTGCTAAAGCTGCGCTT | 680 |
| Ace | ace | F: GGAATGACCGAGAACGATGGC; R: GCTTGATGTTGGCCTGCTTCCG | 616 |
DNAs of all Enterococcus strains were extracted by the boiling method as follows: A loopful of overnight Enterococcus culture was suspended in 300 mL of sterile distilled water; then boiled for 10 minutes and after that centrifuged at 13,000 rpm for 10 minutes. An aliquot of the supernatant (5 mL) was used as the template in a final volume of 25 mL PCR mixture containing: 13 µL PCR buffer, 2 mM MgCl2, 200 nM dNTP, 400 nM of each primer and 0.25 U Taq DNA polymerase (or 20 µL Master mix PCR). Reactions for both mixtures were done on a thermal cycler (Sensoquest, Germany) with an initial denaturation at 95°C for four minutes, 30 cycles of amplification (denaturation at 95°C for 30 seconds, annealing at 52°C for one minute, and extension at 72°C for one minute), and a final extension at 72°C for seven minutes (22, 23).
The multiplex PCR was performed for three different genes (gylE, cylA and esp) using specific primers listed in Table 1. The multiplex PCR mixture was optimized with total volume of 50 µL composed of 25 µL PCR master mix, 0.5 µL of cylA and esp primers and 0.25 µL of gelE, 5 µL of extracted DNA and sterile DNA/RNase distilled water up to 50 µL (22, 23).
To detect ace gene, a separate conventional PCR was performed using specific primers for Enterococcus and following the aforementioned protocol for species-specific PCR; but the annealing temperature was set on 56°C (24, 25). Reactions were performed on thermal cycler with an initial denaturation at 93°C for seven minutes, followed by 30 cycles of denaturation (94°C for one minute), annealing (56°C for one minute), extension (72°C for one minute) and a final extension step at 72°C for 10 minutes.
PCR products were analyzed by electrophoresis on a 3% agarose gel tainted with ethidium bromide and illustrated under UV light. Each PCR assay was supplemented with a negative control, containing all of the reagents without template DNA.
3.5. PCR Primer Design and Amplification of Putative Virulence Genes
The primer sequences and expected size of amplicons for each PCR result are shown in Table 1 (24, 25).
3.6. Statistical Analysis
World health organization (WHO) net, Chi-square and likelihood ratio analysis were carried out using SSPS software version 5.1.
4. Results
Based on the results of various genetic and biochemical tests, out of 149 isolated strains, two species were identified. Enterococcus faecalis (60%) followed by E. faecium (26%) and Enterococcus spp. (14%) were the commonest isolated species (Table 2).
Distribution of Enterococcus spp.
| Code | Organism | Number of Isolates | (%) | Number of Patients | f | m |
|---|---|---|---|---|---|---|
| efa | Enterococcus faecalis | 118 | 60 | 79 | 50 | 68 |
| efm | Enterococcus faecium | 20 | 26 | 13 | 14 | 6 |
| ent | Enterococcus spp. | 11 | 14 | 7 | 8 | 3 |
A total of 149 non-repetitive isolates were analyzed out of which 72 E. faecalis species were isolated from urine (67), pus (1), blood (1), vagina (2) and sterile body fluid (1) samples, and 31 E. faecium species were isolated from pus (3), blood (2) and urine (26) samples.
Clinical samples were identified aseptically in the patients of Milad hospital. The results of these studies showed that the dominant species was E. faecalis followed by E. faecium. All of the 149 Enterococcus isolates were tested by disc diffusion method and the results are shown in Table 3.
As indicated in the Table 3, the highest resistance was observed against gentamicin in all species followed by erythromycin and tetracycline in Enterococcus faecium and E. faecalis, respectively. While minimal resistance to vancomycin was observed too.
Resistance to Different Antibiotics Among Enterococcus faecium, Enterococcus faecalis and Enterococcus spp.
| Antibiotic Name | Breakpoints | Number | %R | %I | %S | % R 95%CI | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| fcla | fcmb | entc | fcl | fcm | ent | fcl | fcm | ent | ||||
| Penicillin G | S ≥ 15 | 149 | 27.1 | 55 | 36.4 | 0 | 0 | 0 | 72.9 | 45 | 63.6 | 24.3 - 39.7 |
| Ampicillin | S ≤ 8; R ≥ 16 | 149 | 26.3 | 65 | 27.3 | 0 | 0 | 0 | 73.7 | 35 | 72.7 | 24.3 - 39.7 |
| Ampicillin | S ≥ 17 | 149 | 24.6 | 50 | 0 | 0 | 0 | 0 | 75.4 | 50 | 100 | 19.5 - 34.1 |
| Gentamicin | 13 - 14 | 149 | 77.1 | 80 | 63.6 | 5.9 | 0 | 0 | 16.9 | 20 | 45.5 | 68.0 - 82.3 |
| Ciprofloxacin | 16 - 20 | 149 | 49.2 | 75 | 54.5 | 33.9 | 15 | 27.3 | 16.9 | 10 | 9.1 | 45.4 - 61.8 |
| Erythromycin | 14 - 22 | 149 | 69.5 | 80 | 54.5 | 27.1 | 15 | 45.5 | 3.4 | 5 | 0 | 61.7 - 76.9 |
| Vancomycin | S ≤ 4; R ≥ 32 | 149 | 28 | 60 | 27.3 | 6.8 | 5 | 9.1 | 65.3 | 35 | 63.6 | 24.9 - 40.4 |
| Vancomycin | 15 - 16 | 149 | 14.4 | 50 | 18.2 | 28 | 15 | 18.2 | 57.6 | 35 | 63.6 | 13.7 - 27.0 |
| Chloramphenicol | 13 - 17 | 149 | 33.9 | 40 | 9.1 | 14.4 | 5 | 27.3 | 51.7 | 55 | 63.6 | 25.6 - 41.1 |
| Tetracycline | 15 - 18 | 149 | 72 | 70 | 63.6 | 6.8 | 5 | 9.1 | 21.2 | 25 | 27.3 | 63.0 - 78.1 |
About 33 strains of VRE, E. faecium, almost half of the resistant strains, were allocated and then, E. faecalis was in the second place. Enterococcus faecalis strains showed the highest resistance to gentamicin. Susceptibility patterns of the isolates are shown in Table 4. Results showed that six isolated strains were resistant to all antibiotics including ampicillin (AMP), gentamicin (GEN), ciprofloxacin (CIP), erythromycin (ERY), vancomycin, chloramphenicol (CHL) and tetracycline (TCY). Meanwhile, more than 34 isolated strains were resistant to five antibiotics including vancomycin, tetracycline, gentamicin, ciprofloxacin and erythromycin. However, 22 strains were resistant to seven categories of various antibiotics. The interesting point was that eight isolates were resistant to only three antibiotics. Results also showed sensitivity to other antibiotics; of the 149 tested Enterococcus strains, 91 were susceptible to vancomycin (MIC ≤ 4 mg/mL), nine were intermediate (MIC = 8 µg/mL for both strains), and 49 were resistant (MIC ≥ 32 mg/mL). The importance of this information lies in the fact that Enterococcus spp. are often potential pathogens of mixed infections for a broad-spectrum antimicrobial agents such as ciprofloxacin and ampicillin.
Resistance Profile of the Isolates to the Studied Antibiotics
| Samples | Resistance Profile |
|---|---|
| 1 | CIP |
| 3 | ERY |
| 1 | ERY, CIP |
| 1 | ERY, CIP, AMP, PEN |
| 2 | ERY, GEN |
| 1 | ERY, GEN, PEN |
| 2 | ERY, GEN, CIP |
| 2 | ERY, GEN, CIP, PEN |
| 1 | ERY, GEN, CIP, AMP |
| 1 | ERY, GEN, CIP, AMP, PEN |
| 1 | GEN |
| 1 | TCY, GEN, CIP, PEN |
| 2 | TCY, ERY |
| 3 | TCY, ERY, GEN |
| 8 | TCY, ERY, GEN, CIP |
| 5 | TCY, ERY, GEN, CIP, PEN |
| 8 | TCY, ERY, GEN, CIP, AMP, PEN |
| 1 | TCY, ERY, CHL, GEN, CIP |
| 3 | TCY, ERY, CHL, GEN, CIP, PEN |
| 1 | TCY, ERY, CHL, GEN, CIP, AMP |
| 3 | TCY, ERY, CHL, GEN, CIP, AMP PEN |
| 1 | VAN, ERY |
| 2 | VAN, ERY, PEN |
| 1 | VAN, ERY,AMP, PEN |
| 1 | VAN, ERY, CIP, AMP, PEN |
| 4 | VAN, ERY, GEN, CIP, AMP, PEN |
| 1 | VAN, ERY, CHL, GEN, CIP, AMP, PEN |
| 2 | VAN, TCY, CIP |
| 5 | VAN, TCY, ERY, GEN, CIP |
| 3 | VAN, TCY, ERY, GEN, CIP, PEN |
| 9 | VAN, TCY, ERY, GEN, CIP, AMP PEN |
| 1 | VAN, TCY, ERY, CHL, CIP, PEN |
| 1 | VAN, TCY, ERY, CHL, CIP, AMP PEN |
| 1 | VAN, TCY, ERY, CHL, GEN, PEN |
| 9 | VAN, TCY, ERY, CHL, GEN, CIP |
| 2 | VAN, TCY, ERY, CHL, GEN, CIP, PEN |
| 1 | VAN, TCY, RY CHL, GEN, CIP, AMP |
| 23 | VAN, TCY, ERY, CHL, GEN, CIP, AMP, PEN |
After the initial identification of different species, the identified species were confirmed by PCR.
Furthermore, PCR was performed on virulence inducing genes. According to the results of the current study shown in Table 5, the esp gene was the most widespread virulence determinant. The gelE was harbored only in 2% of the isolates. The result showed that the ace gene, encoding collagen-binding protein, was found in high frequency among E. faecalis isolates. Forty five isolates of E. faecalis and three isolates of E. faecium had cylA gene.
Distribution of Different Virulence Genes Among Enterococcus Isolates
Agarose Gel Electrophoresis of Amplified cylA, esp and gelE by Multiplex PCR and ace by Simple PCR
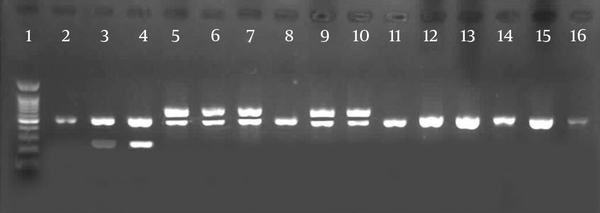
5. Discussion
The current study describes the isolation, biochemical and molecular characterization of intrinsically Enterococcus spp. from urine, blood, pus, stool, fluids and aspirates clinical samples collected from patients admitted to ICU, emergency and surgery rooms of Milad hospital.
Although the prevalence of VRE strains in poultry, food and water were more varied studies also showed that E. faecalis and E. faecium strains are more common (14).
The current study found E. faecalis the predominant detected species with a higher prevalence, as expected. Other studies reported similar results. In comparison to other studies worldwide, resistance of E. faecium to all studied antibiotics was higher than those of E. faecalis. VRE was higher in E. faecium than E. faecalis and E. faecium.
Resistance to multiple classes of antibiotics was common in Enterococcus spp. as observed in this research. The resistance rate to ciprofloxacin and chloramphenicol among E. faecalis was more than that of E. faecium. This may be because of indiscriminate use of these two antimicrobials in human and animal infections and selective pressure or simply the transfer of resistance genes or a combination of both (26).
Maschieto et al. reported that the distribution of Enterococcus spp. isolated from the intestinal tracts of patients referred to a university affiliated hospital in Brazil was E. faecium (34%) followed by E. faecalis (33%), E. gallinarum (23.7%), E. casseliflavus (5.2%), E. avium (1%) and E. hirae (1.2%) (27).
These results are consistent with other studies conducted in Iran and in environmental samples (24). This may be due to the ability of resistance to antimicrobial agents and because of that the bacteria is one of the most opportunistic pathogens. Prolonged hospital stay, inappropriate use of antibiotics such as cephalosporin and vancomycin, use of antimicrobial growth promoters such as avoparcin in animal food, organ transplantation, using metronidazole, surgery, diabetes and leukemia for some reasons can be a predisposing factor in colonization or infection with these microorganisms. According to the evidences, VRE can be transmitted horizontally from human to human and animal to human (26).
In the current study, 32.2% of the isolates were resistant to vancomycin. Compared with other studies conducted worldwide, this amount is different and variable. The study by Gambarotto et al. showed that the prevalence of VRE in hematology patients was about 37% (25).
Kuhn et al. showed that the prevalence of VRE isolated from animals, humans and the environment in different European regions was in the range of 8% - 11% (16).
The study by Martinez et al. showed significant relationship between antibiotic usage (vancomycin, cephalosporin, metronidazole and quinolones) and emergence of VRE (28).
In the United States, VRE isolates are restricted to hospitalized patients, whereas in European countries VRE are isolated from different environments (29). The prevalence of vancomycin resistant strains in Europe and the United States is in association with the excessive use of glycopeptide antibiotics in health centers due to the isolation of these bacteria from human gut and colonization in the gastrointestinal tract or transferring the resistance genes to microorganisms of digestive tract; while the transition and use of growth promoters such as avoparcin as a food supplement for livestock and poultry (26).
The release of the microorganism takes place from patient to patient via the contaminated hands of personnel. The bacteria remain alive on the hands for 30 minutes. It is difficult to eliminate even a VRE from a hospital (30).
About 53.7% of the strains were resistant to ciprofloxacin and 32.2% were resistant to vancomycin. This antibiotic is widely used due to the success in the treatment of urinary tract infections. In 2000, a report in Greece showed that E. faecium had grater resistance to antibiotics (three times more) than Enterococcus faecalis (21).
Resistance to erythromycin in the current study was 69% of the reported strains that was due to the uncontrolled use of these antibiotics in Iran. Erythromycin is an antibiotic used in the food industry that affects resistance in Enterococcus spp. (18). Stobberingh et al. showed that resistance to vancomycin in clinical and environmental samples was 12% and 12%, respectively (31).
In general, the rate of VRE strains resistant to gentamicin was reported 86.6% and about 76%. As predicted, most of E. faecium strains were resistant to gentamicin. However, Khan et al. reported that resistance to gentamicin was about 96% in America (23).
In the current study, 71.1% of the strains were resistant to tetracycline and 32.2% were resistant to vancomycin. The overall level of resistance to tetracycline in the world is highly variable. In this study, the lowest resistance belonged to vancomycin.
On the other hand, Enterococcus species have the ability to reproduce and survive in soil and water. Therefore, a lot of attention should be paid to prevent transmission of microorganisms in the nature.
In the present study, resistance to penicillin was observed in 27.1% of E. faecalis and 55% of E. faecium (Table 3).
Antibiotic resistance alone cannot explain the virulence of Enterococcus spp. In order to become pathogenic, they need to express virulence traits associated with adhesion, translocation and evasion of immune responses and cause pathological changes (32).
In the present study, ace, esp, cylA and gelE genes were detected with 39.5%, 61.7%, 32.2% and 2% in isolated strains, respectively. In general, the incidence of these virulence traits was lower among E. faecium strains than E. faecalis strains. The four examined genes were more prevalent in Enterococcus faecalis than in Enterococcus faecium.
According to the results of the current study, E. faecium strains isolated from clinical samples had lower potential pathogenicity than those of the E. faecalis. However, virulence genes in E. faecalis isolates were more variable.
Authors observed a considerable number of E. faecalis harboring esp compared with cylA and gelE genes. Shankar et al. showed that 29% of the blood isolates and 42% of endocarditis E. faecalis isolates were positive for the esp gene, while only 3% of species isolated from stool showed this trait (33).
In the present investigation, the esp gene, which encodes enterococcal surface protein, was found in high frequency among E. faecalis strains (Table 5). A high incidence of this gene in E. faecalis was reported in previous studies (34).
The contribution of the surface protein Esp to colonization and insistence of E. faecalis in urinary tract infections was observed in an animal model (33).
Franz et al. previously reported that the presence of virulence factors is a strain specific character (35). Similarly, a high distribution of the gelE gene in E. faecalis was reported by Mannu et al. (36).
Only 20% of the E. faecium strains but 44.9% of the E. faecalis strains produced ace (Table 5).
Conversely, a study on food and medical isolates showed the incidence of esp in clinical E. faecium isolates is more than that of E.faecalis (32).
Vancomycin resistance was not associated with more virulent strains in the current study. In fact, according to Giridhara Upadhyaya et al. (37) there is insignificant difference in virulence factors, ability to cause infection or vancomycin susceptibility among Enterococcus isolates.
In the current study, gelE significantly enriched in E. faecalis isolates in comparison with E. faecium and may involve in the creation of a urinary tract infection (P < 0.001). The obtained results showed that, gelE-positive E. faecalis isolates had less frequency in clinical isolates and not found in any of the E. faecium isolates. Likewise, previous studies on E. faecalis demonstrated high expression of this gene among the isolates (8, 34).
5.1. Conclusion
There were some limitations in the current study such as small sample size, short duration of study, and inability to detect different strains of Enterococcus spp. The results of the study indicated that more research is needed to characterize molecular and cellular interactions between the host and Enterococcus isolates which lead to intra-species genetic transfer and virulence factors. The identification of virulence factors associated with invasiveness and disease severity is an important subject for research.
A better understanding of the role of the virulence factors of Enterococcus spp. in nosocomial infections may help to improve new strategies to prevent or reduce the infection by this species.
Acknowledgements
References
-
1.
Magi G, Capretti R, Paoletti C, Pietrella M, Ferrante L, Biavasco F, et al. Presence of a vanA-carrying pheromone response plasmid (pBRG1) in a clinical isolate of Enterococcus faecium. Antimicrob Agents Chemother. 2003;47(5):1571-6. [PubMed ID: 12709324].
-
2.
Heidari H, Emaneini M, Dabiri H, Jabalameli F. Virulence factors, antimicrobial resistance pattern and molecular analysis of Enterococcal strains isolated from burn patients. Microb Pathog. 2016;90:93-7. [PubMed ID: 26620079].
-
3.
Talebi M, Rahimi F, Katouli M, Kühn I, Möllby R, Eshraghi S, et al. Prevalence and antimicrobial resistance of enterococcal species in sewage treatment plants in Iran. Water Air Soil Pollut. 2007;185(1-4):111-9.
-
4.
Laukova A, Kandricakova A, Scerbova J, Strompfova V. Enterococci isolated from farm ostriches and their relation to enterocins. Folia microbiol. 2015:1-7.
-
5.
Marshall CG, Lessard IA, Park I, Wright GD. Glycopeptide antibiotic resistance genes in glycopeptide-producing organisms. Antimicrob Agents Chemother. 1998;42(9):2215-20. [PubMed ID: 9736537].
-
6.
Morrison AJ, Wenzel RP. Nosocomial urinary tract infections due to enterococcus. Ten years' experience at a university hospital. Arch Intern Med. 1986;146(8):1549-51. [PubMed ID: 3729635].
-
7.
Fisher K, Phillips C. The ecology, epidemiology and virulence of Enterococcus. Microbiology. 2009;155(Pt 6):1749-57. [PubMed ID: 19383684]. https://doi.org/10.1099/mic.0.026385-0.
-
8.
Flores RM, Ross TW. Vancomycin-Resistant Enterococci: Approach to Treatment and Control. Cancer Control. 1996;3(1):66-72. [PubMed ID: 10825278].
-
9.
Kandricakova A, Laukova A, Strompfova V. Characteristic and susceptibility to enterocins of enterococci in pheasants possessing virulence factor genes. Pol J Vet Sci. 2015;18(3):507-14. [PubMed ID: 26618582]. https://doi.org/10.1515/pjvs-2015-0066.
-
10.
Eisner A, Feierl G, Gorkiewicz G, Dieber F, Kessler HH, Marth E, et al. High prevalence of VanA-type vancomycin-resistant Enterococci in Austrian poultry. Appl Environ Microbiol. 2005;71(10):6407-9. [PubMed ID: 16204568]. https://doi.org/10.1128/AEM.71.10.6407-6409.2005.
-
11.
Macovei L, Zurek L. Ecology of antibiotic resistance genes: characterization of enterococci from houseflies collected in food settings. Appl Environ Microbiol. 2006;72(6):4028-35. [PubMed ID: 16751512]. https://doi.org/10.1128/AEM.00034-06.
-
12.
Arias CA, Contreras GA, Murray BE. Management of multidrug-resistant enterococcal infections. Clin Microbiol Infect. 2010;16(6):555-62. [PubMed ID: 20569266]. https://doi.org/10.1111/j.1469-0691.2010.03214.x.
-
13.
Sharifi Y, Hasani A, Ghotaslou R, Naghili B, Aghazadeh M, Milani M, et al. Virulence and antimicrobial resistance in enterococci isolated from urinary tract infections. Adv Pharm Bull. 2013;3(1):197-201. [PubMed ID: 24312835]. https://doi.org/10.5681/apb.2013.032.
-
14.
Talebi M, Rahimi F, Katouli M, Mollby R, Pourshafie MR. Epidemiological link between wastewater and human vancomycin-resistant Enterococcus faecium isolates. Curr Microbiol. 2008;56(5):468-73. [PubMed ID: 18293035]. https://doi.org/10.1007/s00284-008-9113-0.
-
15.
Tendolkar PM, Baghdayan AS, Shankar N. Pathogenic enterococci: new developments in the 21st century. Cell Mol Life Sci. 2003;60(12):2622-36. [PubMed ID: 14685687]. https://doi.org/10.1007/s00018-003-3138-0.
-
16.
Kuhn I, Iversen A, Finn M, Greko C, Burman LG, Blanch AR, et al. Occurrence and relatedness of vancomycin-resistant enterococci in animals, humans, and the environment in different European regions. Appl Environ Microbiol. 2005;71(9):5383-90. [PubMed ID: 16151128]. https://doi.org/10.1128/AEM.71.9.5383-5390.2005.
-
17.
Kayaoglu G, Orstavik D. Virulence factors of Enterococcus faecalis: relationship to endodontic disease. Crit Rev Oral Biol Med. 2004;15(5):308-20. [PubMed ID: 15470268].
-
18.
Cetinkaya Y, Falk P, Mayhall CG. Vancomycin-resistant enterococci. Clin Microbiol Rev. 2000;13(4):686-707. [PubMed ID: 11023964].
-
19.
Dupre I, Zanetti S, Schito AM, Fadda G, Sechi LA. Incidence of virulence determinants in clinical Enterococcus faecium and Enterococcus faecalis isolates collected in Sardinia (Italy). J Med Microbiol. 2003;52(Pt 6):491-8. [PubMed ID: 12748268]. https://doi.org/10.1099/jmm.0.05038-0.
-
20.
Novais C, Coque TM, Ferreira H, Sousa JC, Peixe L. Environmental contamination with vancomycin-resistant enterococci from hospital sewage in Portugal. Appl Environ Microbiol. 2005;71(6):3364-8. [PubMed ID: 15933043]. https://doi.org/10.1128/AEM.71.6.3364-3368.2005.
-
21.
Platsouka ED, Dimopoulou H, Miriagou V, Paniara O. The first clinical isolates of Enterococcus faecium with the VanA phenotype in a tertiary Greek hospital. J Antimicrob Chemother. 2000;46(6):1039-40. [PubMed ID: 11102430].
-
22.
Vankerckhoven V, Van Autgaerden T, Vael C, Lammens C, Chapelle S, Rossi R, et al. Development of a multiplex PCR for the detection of asa1, gelE, cylA, esp, and hyl genes in enterococci and survey for virulence determinants among European hospital isolates of Enterococcus faecium. J Clin Microbiol. 2004;42(10):4473-9. [PubMed ID: 15472296]. https://doi.org/10.1128/JCM.42.10.4473-4479.2004.
-
23.
Khan SA, Nawaz MS, Khan AA, Hopper SL, Jones RA, Cerniglia CE. Molecular characterization of multidrug-resistant Enterococcus spp. from poultry and dairy farms: detection of virulence and vancomycin resistance gene markers by PCR. Mol Cell Probes. 2005;19(1):27-34. [PubMed ID: 15652217]. https://doi.org/10.1016/j.mcp.2004.09.001.
-
24.
Rahimi F, Talebi M, Saifi M, Pourshafie MR. Distribution of enterococcal species and detection of vancomycin resistance genes by multiplex PCR in Tehran sewage. Iran Biomed J. 2007;11(3):161-7. [PubMed ID: 18051776].
-
25.
Gambarotto K, Ploy MC, Turlure P, Grelaud C, Martin C, Bordessoule D, et al. Prevalence of vancomycin-resistant enterococci in fecal samples from hospitalized patients and nonhospitalized controls in a cattle-rearing area of France. J Clin Microbiol. 2000;38(2):620-4. [PubMed ID: 10655356].
-
26.
Arbabi L, Vandyoudefi J, Bouzari M, Rahimi F, Rahimi F. Antibiotic susceptibility pattern among vancomycin resistant enterococci isolated from meat and fecal samples in tehran livestock husbandries. 2010.
-
27.
Maschieto A, Martinez R, Palazzo IC, Darini AL. Antimicrobial resistance of Enterococcus sp. isolated from the intestinal tract of patients from a university hospital in Brazil. Mem Inst Oswaldo Cruz. 2004;99(7):763-7. [PubMed ID: 15654436].
-
28.
Martinez JA, Ruthazer R, Hansjosten K, Barefoot L, Snydman DR. Role of environmental contamination as a risk factor for acquisition of vancomycin-resistant enterococci in patients treated in a medical intensive care unit. Arch Intern Med. 2003;163(16):1905-12. [PubMed ID: 12963563]. https://doi.org/10.1001/archinte.163.16.1905.
-
29.
Goossens H. The epidemiology of vancomycin-resistant enterococci. Curr Opin Infect Dis. 1999;12(6):537-41. [PubMed ID: 17035818].
-
30.
Gikas A, Christidou A, Scoulica E, Nikolaidis P, Skoutelis A, Levidiotou S, et al. Epidemiology and molecular analysis of intestinal colonization by vancomycin-resistant enterococci in greek hospitals. J Clin Microbiol. 2005;43(11):5796-9. [PubMed ID: 16272524]. https://doi.org/10.1128/JCM.43.11.5796-5799.2005.
-
31.
Stobberingh E, van den Bogaard A, London N, Driessen C, Top J, Willems R. Enterococci with glycopeptide resistance in turkeys, turkey farmers, turkey slaughterers, and (sub)urban residents in the south of The Netherlands: evidence for transmission of vancomycin resistance from animals to humans? Antimicrob Agents Chemother. 1999;43(9):2215-21. [PubMed ID: 10471567].
-
32.
Eaton TJ, Gasson MJ. Molecular screening of Enterococcus virulence determinants and potential for genetic exchange between food and medical isolates. Appl Environ Microbiol. 2001;67(4):1628-35. [PubMed ID: 11282615]. https://doi.org/10.1128/AEM.67.4.1628-1635.2001.
-
33.
Shankar V, Baghdayan AS, Huycke MM, Lindahl G, Gilmore MS. Infection-derived Enterococcus faecalis strains are enriched in esp, a gene encoding a novel surface protein. Infect Immun. 1999;67(1):193-200. [PubMed ID: 9864215].
-
34.
Sharifi Y, Hasani A, Ghotaslou R, Varshochi M, Hasani A, Aghazadeh M, et al. Survey of Virulence Determinants among Vancomycin Resistant Enterococcus faecalis and Enterococcus faecium Isolated from Clinical Specimens of Hospitalized Patients of North west of Iran. Open Microbiol J. 2012;6:34-9. [PubMed ID: 22582098]. https://doi.org/10.2174/1874285801206010034.
-
35.
Franz CM, Muscholl-Silberhorn AB, Yousif NM, Vancanneyt M, Swings J, Holzapfel WH. Incidence of virulence factors and antibiotic resistance among Enterococci isolated from food. Appl Environ Microbiol. 2001;67(9):4385-9. [PubMed ID: 11526054].
-
36.
Mannu L, Paba A, Daga E, Comunian R, Zanetti S, Dupre I, et al. Comparison of the incidence of virulence determinants and antibiotic resistance between Enterococcus faecium strains of dairy, animal and clinical origin. Int J f food microbiol. 2003;88(2):291-304.
-
37.
Giridhara Upadhyaya PM, Ravikumar KL, Umapathy BL. Review of virulence factors of enterococcus: an emerging nosocomial pathogen. Indian J Med Microbiol. 2009;27(4):301-5. [PubMed ID: 19736397]. https://doi.org/10.4103/0255-0857.55437.