Abstract
Background:
Nontuberculous mycobacteria (NTM) are opportunist pathogens, which can cause pulmonary and non-pulmonary diseases.Objectives:
This study aimed at investigating the frequency of NTM at a teaching hospital of Tehran, Iran.Methods:
Clinical samples from patients with signs and symptoms of TB were collected from March 2016 to July 2017. Specimens were investigated using conventional methods and the GeneXpert MTB/RIF system. Phenotypic, polymerase chain reaction (PCR)-restriction enzyme analysis (PRA) and sequencing methods were used to identify the mycobacterial species.Results:
Of 230 culture positive samples, 12 (5.2%) were NTM, according to conventional and molecular approaches. Mycobacterium simiae (6, 50.0%), M. fortuitum (4, 33.3%), M. intracellulare (1, 8.3%) and M. kansasii (1, 8.3%) were the isolated NTM species.Conclusions:
The current study strongly highlights the need to design strategies for surveillance, monitoring, and management of NTM cases.Keywords
1. Background
Non-Tuberculosis Mycobacteria (NTM) are environmental organisms that can cause opportunistic infections in humans. While many NTM species are considered as free-living saprophytes, an increasing number have been reported as opportunistic pathogens, which are capable of causing serious diseases in humans (1, 2). In the last decade, the prevalence of NTM has been increasing worldwide and NTM accounts for an increasing proportion of mycobacterial diseases (2-4). Furthermore, NTM infections are similar to tuberculosis (TB) in their clinical and microbiological presentation and may cause a clinical dilemma with regards to therapy for infected patients (5). In Iran and most other developing countries, in which TB is an endemic disease, diagnosis of TB is difficult and clinicians mostly rely on conventional tests (i.e. tuberculin skin testing, chest radiography, and microscopy examination), a practice that may not distinguish NTM from TB (4, 6). A recent report from Iran found that 30% of patients with NTM infections were wrongly diagnosed as MDR-TB (6).
2. Objectives
Given the above facts, this study aimed at reporting the prevalence of NTM among TB suspects, referred to a teaching hospital in Tehran, Iran.
3. Methods
3.1. Samples and Setting
A total of 230 clinical isolates from TB suspected cases that were referred to Baqiyatallah hospital from March 2016 to July 2017 were included in this study. This hospital has well-equipped biosafety level III laboratory facilities and standard safety precautions were followed for specimen processing. The ethics committee of Baqiyatallah hospital approved the study and all the patients signed an informed consent.
3.2. Isolation and Identification of Mycobacteria
Obtained specimens were processed using Petroff’s method (7). The obtained sediments were stained with Ziehl-Neelsen and were cultured in Lowenstein-Jensen medium (7). Culture media was examined twice per week for growth rate, colony morphology, and pigmentation, according to standard procedures (8).
3.3. Identification of M. tuberculosis
GeneXpert assay was performed for assignment of isolates to M. tuberculosis, as described previously (9, 10). Briefly, sample reagent was added at a 2:1 ratio to clinical specimens and then the mixture was added to the Xpert MTB/RIF cartridge to perform the assay in the Genexpert instrument. The automatically generated results were read after 90 minutes.
3.4. Identification of NTM Species
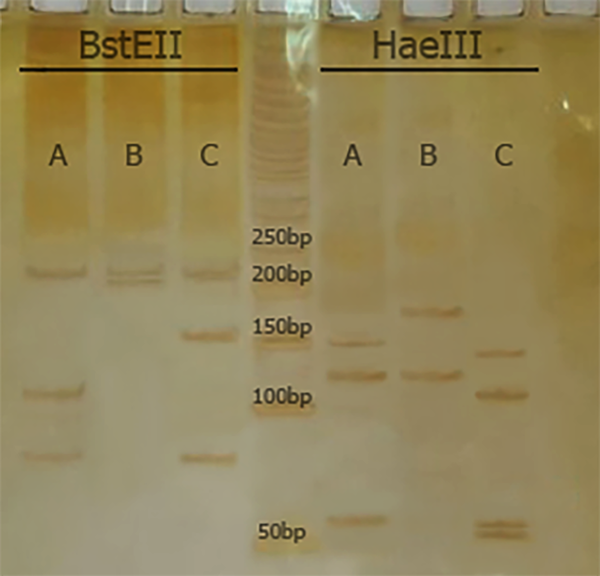
Polymerase Chain Reaction restriction analysis (PRA) and hsp65 gene sequencing were used to speciate NTM. For PRA, a set of primers (Tb11 and Tb12) was used to amplify a 440-bp fragment (Tb11: 5’-ACCAACGATGGTGTGTCCAT-3’, and Tb12: 5’-CTTGTCGAACCGCATACCCT-3’). The Hae III and Bst II restriction enzymes were used to digest the amplified products and electrophoresis was performed on 8% polyacrylamide gel (11). The algorithm proposed by Roth et al. was used for species identification (12).
For sequencing, the amplified PCR products of hsp65 gene for each isolate were sequenced using specific primers, as described previously (11).
4. Results
4.1. NTM Isolation
Of 230 culture positive TB isolates, 12 isolates (5.2%) were NTM. The remaining, 218 isolates were confirmed as M. tuberculosis using the GeneXpert method.
4.2. PRA and hsp65 Sequencing
According to PRA and hsp65 sequencing, M. simiae (6, 50.0%), M. fortuitum (4, 33.3%), M. intracellulare (1, 8.3%), and M. kansasii (1, 8.3%) were the most commonly isolated NTM, respectively (Figure 1 and Table 1). Clinical isolates were confidently identified by hsp65 gene sequencing.
Comparison of BstEII and HaeIII RFLP patterns from isolates of M. tuberculosis (A: 235/120/85 and 150/130/70), M. simiae (B: 235/210 and 185/130) and M. fortuitum (C: 235/120/85 and 145/120/60/55) isolates from clinical samples.
Results of NTM Identification by Phenotypic and Genotypic Tests
| Numbers of Isolates | Phenotypic Tests | Patterns by hsp65-PRA | Identification by PRA | hsp65 Sequencing | |
|---|---|---|---|---|---|
| Bst E II | Hae III | ||||
| 6 | Mycobacterium sp. | 235/210 | 185/130 | M. simiae | M. simiae |
| 4 | Mycobacterium sp. | 235/120/85 | 145/120/60/55 | M. fortuitum | M. fortuitum |
| 1 | Mycobacterium sp. | 235/120/100 | 145/130/60 | M. intracellulare | M. intracellulare |
| 1 | Mycobacterium sp. | 235/210 | 130/105/80 | M. kansasii | M. kansasii |
5. Discussion
In Iran, diseases caused by NTM appear to be increasing and represent an important public-health threat. Although knowledge about NTM diseases seems to be improved, yet reliable diagnosis of NTM still remains problematic.
According to the current study, 5.2% of isolates, which were obtained from TB-suspected patients were positive for NTM. Several reports suggest that there is an increasing number of NTM worldwide (13-19). The current researchers have recently shown a relatively high prevalence of NTM infections (10.2%) among culture-positive cases of TB, which emphasizes on the role of NTM in public health (4).
Mistaking patients with NTM for TB is a serious error (6). So far, all NTM are acid-fast; clinical manifestations of many NTM are often indistinguishable from TB and disease caused by NTM, usually does not respond to anti-TB drugs (6, 20). Thus, accurate diagnosis of mycobacterial species is very important and can facilitate the prevention and control of NTM infections.
Mycobacterium simiae was the most common NTM species from investigated isolates in the current study. This was similar to previous studies, which were conducted in Iran (21-23). Mycobacterium simiae is an endemic NTM in Iran and its clinical and microbiological findings are often indistinguishable from TB (21, 22). Thus, the relatively high prevalence of M. simiae may cause serious problems for TB control strategies in Iran.
Mycobacterium fortuitum and M. kansasii were also among the most common NTM species, which were isolated from suspected TB patients. Similarly, Hashemi-Shahraki et al. found that M. fortuitum and M. kansasii were the most common cause of NTM diseases in Ahvaz (a city located in the south of Iran) (19). They evaluated 92 isolates from patients with confirmed NTM diseases and concluded that PRA and sequencing are reliable methods for elucidating taxonomic data and species identification of Mycobacterium isolates (19).
The first limitation of the current study was the relatively small clinical sample size, thus, this research have underestimated the true prevalence of NTM. Secondly, although the current patients were likely to have pulmonary NTM disease, yet the researchers were unable to completely rule out the possibility of NTM contamination of the positive cultures. In conclusion, the relatively high isolation of NTM strongly highlights the need to make strategies for surveillance, monitoring and management of NTM cases.
References
-
1.
Novosad S, Henkle E, Winthrop KL. The Challenge of Pulmonary Nontuberculous Mycobacterial Infection. Curr Pulmonol Rep. 2015;4(3):152-61. [PubMed ID: 26877911]. [PubMed Central ID: PMC4749270]. https://doi.org/10.1007/s13665-015-0119-3.
-
2.
Winthrop KL, McNelley E, Kendall B, Marshall-Olson A, Morris C, Cassidy M, et al. Pulmonary nontuberculous mycobacterial disease prevalence and clinical features: an emerging public health disease. Am J Respir Crit Care Med. 2010;182(7):977-82. [PubMed ID: 20508209]. https://doi.org/10.1164/rccm.201003-0503OC.
-
3.
Mirsaeidi M, Machado RF, Garcia JG, Schraufnagel DE. Nontuberculous mycobacterial disease mortality in the United States, 1999-2010: a population-based comparative study. PLoS One. 2014;9(3). e91879. [PubMed ID: 24632814]. [PubMed Central ID: PMC3954860]. https://doi.org/10.1371/journal.pone.0091879.
-
4.
Nasiri MJ, Dabiri H, Darban-Sarokhalil D, Hashemi Shahraki A. Prevalence of Non-Tuberculosis Mycobacterial Infections among Tuberculosis Suspects in Iran: Systematic Review and Meta-Analysis. PLoS One. 2015;10(6). e0129073. [PubMed ID: 26052701]. [PubMed Central ID: PMC4460155]. https://doi.org/10.1371/journal.pone.0129073.
-
5.
Gopinath K, Singh S. Non-tuberculous mycobacteria in TB-endemic countries: are we neglecting the danger? PLoS Negl Trop Dis. 2010;4(4). e615. [PubMed ID: 20436962]. [PubMed Central ID: PMC2860495]. https://doi.org/10.1371/journal.pntd.0000615.
-
6.
Shahraki AH, Heidarieh P, Bostanabad SZ, Khosravi AD, Hashemzadeh M, Khandan S, et al. "Multidrug-resistant tuberculosis" may be nontuberculous mycobacteria. Eur J Intern Med. 2015;26(4):279-84. [PubMed ID: 25784643]. [PubMed Central ID: PMC4414892]. https://doi.org/10.1016/j.ejim.2015.03.001.
-
7.
Nasiri MJ, Rezaei F, Zamani S, Darban-Sarokhalil D, Fooladi AA, Shojaei H, et al. Drug resistance pattern of Mycobacterium tuberculosis isolates from patients of five provinces of Iran. Asian Pac J Trop Med. 2014;7(3):193-6. [PubMed ID: 24507638]. https://doi.org/10.1016/S1995-7645(14)60019-5.
-
8.
Kent PT, Kubica GP; Centers for Disease Control. Public Health Mycobacteriology: A Guide for the Level III Laboratory. U.S. Department of Health and Human Services, Public Health Service, Centers for Disease Control; 1985.
-
9.
Boehme CC, Nabeta P, Hillemann D, Nicol MP, Shenai S, Krapp F, et al. Rapid molecular detection of tuberculosis and rifampin resistance. N Engl J Med. 2010;363(11):1005-15. [PubMed ID: 20825313]. [PubMed Central ID: PMC2947799]. https://doi.org/10.1056/NEJMoa0907847.
-
10.
Helb D, Jones M, Story E, Boehme C, Wallace E, Ho K, et al. Rapid detection of Mycobacterium tuberculosis and rifampin resistance by use of on-demand, near-patient technology. J Clin Microbiol. 2010;48(1):229-37. [PubMed ID: 19864480]. [PubMed Central ID: PMC2812290]. https://doi.org/10.1128/JCM.01463-09.
-
11.
Telenti A, Marchesi F, Balz M, Bally F, Bottger EC, Bodmer T. Rapid identification of mycobacteria to the species level by polymerase chain reaction and restriction enzyme analysis. J Clin Microbiol. 1993;31(2):175-8. [PubMed ID: 8381805]. [PubMed Central ID: PMC262730].
-
12.
Roth A, Reischl U, Streubel A, Naumann L, Kroppenstedt RM, Habicht M, et al. Novel diagnostic algorithm for identification of mycobacteria using genus-specific amplification of the 16S-23S rRNA gene spacer and restriction endonucleases. J Clin Microbiol. 2000;38(3):1094-104. [PubMed ID: 10699003]. [PubMed Central ID: PMC86348].
-
13.
Shafipour M, Ghane M, Alang SR, Livani S, Javid N, Shakeri F. Non tuberculosis Mycobacteria isolated from tuberculosis patients in Golestan province, North of Iran. Ann Biol Res. 2013;4:133-7.
-
14.
Velayati AA, Farnia P, Mozafari M, Malekshahian D, Seif S, Rahideh S, et al. Molecular epidemiology of nontuberculous mycobacteria isolates from clinical and environmental sources of a metropolitan city. PLoS One. 2014;9(12). e114428. [PubMed ID: 25485795]. [PubMed Central ID: PMC4259318]. https://doi.org/10.1371/journal.pone.0114428.
-
15.
Khosravi AD, Seghatoleslami S, Hashemzadeh M. Application of PCR-based fingerprinting for detection of nontuberculous mycobacteria among patients referred to tuberculosis reference center of Khuzestan Province, Iran. Res J Microbiol. 2009;4(4):143-9. https://doi.org/10.3923/jm.2009.143.149.
-
16.
Moore JE, Kruijshaar ME, Ormerod LP, Drobniewski F, Abubakar I. Increasing reports of non-tuberculous mycobacteria in England, Wales and Northern Ireland, 1995-2006. BMC Public Health. 2010;10:612. [PubMed ID: 20950421]. [PubMed Central ID: PMC2964631]. https://doi.org/10.1186/1471-2458-10-612.
-
17.
Donohue MJ, Mistry JH, Donohue JM, O'Connell K, King D, Byran J, et al. Increased Frequency of Nontuberculous Mycobacteria Detection at Potable Water Taps within the United States. Environ Sci Technol. 2015;49(10):6127-33. [PubMed ID: 25902261]. https://doi.org/10.1021/acs.est.5b00496.
-
18.
Prevots DR, Marras TK. Epidemiology of human pulmonary infection with nontuberculous mycobacteria: a review. Clin Chest Med. 2015;36(1):13-34. [PubMed ID: 25676516]. [PubMed Central ID: PMC4332564]. https://doi.org/10.1016/j.ccm.2014.10.002.
-
19.
Hashemi-Shahraki A, Bostanabad SZ, Heidarieh P, Titov LP, Khosravi AD, Sheikhi N, et al. Species spectrum of nontuberculous mycobacteria isolated from suspected tuberculosis patients, identification by multi locus sequence analysis. Infect Genet Evol. 2013;20:312-24. [PubMed ID: 24070831]. https://doi.org/10.1016/j.meegid.2013.08.027.
-
20.
Maiga M, Siddiqui S, Diallo S, Diarra B, Traore B, Shea YR, et al. Failure to recognize nontuberculous mycobacteria leads to misdiagnosis of chronic pulmonary tuberculosis. PLoS One. 2012;7(5). e36902. [PubMed ID: 22615839]. [PubMed Central ID: PMC3353983]. https://doi.org/10.1371/journal.pone.0036902.
-
21.
Hashemi-Shahraki A, Darban-Sarokhalil D, Heidarieh P, Feizabadi MM, Deshmir-Salameh S, Khazaee S, et al. Mycobacterium simiae: a possible emerging pathogen in Iran. Jpn J Infect Dis. 2013;66(6):475-9. [PubMed ID: 24270133].
-
22.
Baghaei P, Tabarsi P, Farnia P, Marjani M, Sheikholeslami FM, Chitsaz M, et al. Pulmonary disease caused by Mycobacterium simiae in Iran's national referral center for tuberculosis. J Infect Dev Ctries. 2012;6(1):23-8. [PubMed ID: 22240424].
-
23.
Shojaei H, Heidarieh P, Hashemi A, Feizabadi MM, Daei Naser A. Species identification of neglected nontuberculous mycobacteria in a developing country. Jpn J Infect Dis. 2011;64(4):265-71. [PubMed ID: 21788699].