Abstract
Background:
Accurate speciation of the clinical yeast isolates is essential to a targeted treatment.Objectives:
The aim of this study was to identify and determine the frequency of uncommon and rare yeast species causing fungal infections that may be misidentified.Methods:
During a 10-month period, yeast isolates collected from patients referring to or hospitalized in some educational hospitals in Tehran, Iran, were included in this study. In addition to conventional methods such as direct microscopy and culture on CHROMagar Candida, molecular methods including PCR-RFLP and ITS-sequencing were used for the accurate identification of the yeast strains.Results:
Among 930 yeast isolates recovered from normally sterile and other clinical specimens, a total number of 27 strains were identified as uncommon Candida species and three were identified as rare non-Candida species yeasts. They included C. kefyr (n = 12), C. lusitaniae (n = 8), C. intermedia (n = 3), C. orthopsilosis, C. guilliermondii, and Trichosporon asahii (each n = 2), and Magnusiomyces capitatus (n = 1).Conclusions:
The isolation of less common or rare yeast species, which can cause a variety of infections from superficial to systemic infections, is increasingly reported. Since this uncommon yeast species may exhibit low susceptibility to some antifungal agents, the use of reliable methods for accurate screening of these yeasts is necessary.Keywords
1. Background
Yeasts, which once were considered as non-pathogenic organisms or as normal microbiota of the skin, mucosal oral cavity, gastrointestinal tract, and vagina of the healthy human body, are nowadays recognized as the important causes of life-threating disseminated infections in both immunocompromised and immunocompetent individuals (1). Candida species are the most common yeasts responsible for a wide range of infections from mucocutaneous candidiasis to candidemia and deep tissue infections (2-4). While more than 90% of invasive infections are caused by C. albicans, C. glabrata, C. parapsilosis, C. tropicalis, and C. krusei (5), some rare Candida species, Trichosporon, Geotrichum, and Rhodotorula species have been emerged as opportunistic pathogens causing serious lethal infections (6). The target hosts are immunocompromised patients, including those receiving broad-spectrum antimicrobial agents and organ transplant or prosthetic device recipients, who have had hyperalimentation or indwelling intravenous and urinary catheter insertions (7, 8). Treatments of these infections can be problematic due to the lack of enough information and common resistance to some antifungals (9). Furthermore, the spectrum of diseases caused by the members of the genus in different hosts is incompletely characterized and the clinical manifestation of infections are usually indistinguishable (6, 10). In addition, these fungi may colonize the skin and contaminate the blood cultures; therefore, the significance and interpretation of recovering yeasts remain challenging (6, 10, 11). The correct identification of yeasts at the species level is crucial in order for appropriate, early, and effective antifungal therapy.
2. Objectives
Since the treatment of yeast infections is challenging, owing to the misidentifications of causative agents and due to the resistance of some strains to antifungal agents, the aim of the present study was to identify and determine the frequency of uncommon yeast isolates among patients with suspected superficial or deep-seated fungal infections.
3. Methods
This cross-sectional study was carried out on the samples obtained from patients referring to teaching centers and hospitals, including medical mycology laboratory at the School of Public Health, Tehran University of Medical Sciences, Children’s Medical Centre, Imam Khomeini Hospital, Shariati Hospital, Baghiyatallah Hospital, and Ali-Asghar Hospital, all in Tehran, Iran, between August 2015 and July 2016. Yeast strains were recovered from a variety of specimens including skin and nail scrapings, oral cavity and other mucous membranes, throat swabs, urine, wound discharges, sputum and tracheal tube secretion, BAL, CSF, blood, stool, and biopsies. The unsterile clinical samples were plated on Sabouraud dextrose agar (SDA) with 50 mg chloramphenicol/L (Difco, Detroit, MI, USA) and incubated at 35°C for 10 days. The sterile specimens were inoculated into blood culture bottles (BACTEC Peds Plus/F Culture Vials, Ireland) and were processed according to routine practice using the automated blood culture system BACTEC 9120 (Becton Dickinson, USA) with an incubation period of five days. A positive culture was sub-cultured on SDA and the purity of the isolate was assessed by sub-culturing on CHROMagar Candida medium (CHROMagar Paris, France), which distinguishes mixed isolates based on colony color produced by the yeasts. After incubation at 35°C for 48 h, C. albicans, C. tropicalis, and C. krusei were identified and excluded based on their colony color. Other cream-to-pink or purple colonies were subjected to the PCR-RFLP method as described previously (7). Briefly, DNA was extracted from colonies by the boiling method (12); the entire ITS1-5.8SrDNA-ITS2 region was PCR-amplified with ITS1 (5′-TCC GTA GGT GAA CCT GCG G-3′) and ITS4 (5′-TCC TCC GCT TAT TGA TAT GC-3′), followed by restriction digestion of the products by the enzyme MspI (Fermentas, Vilnius, Lithuania), electrophoresed on agarose gels, and visualized under ultraviolet light. The strains with no specific digestion pattern were subjected to the sequencing of the entire ITS region by forward primer and analyzed as described before (13, 14). Briefly, the ITS1-5.8S-ITS2 regions of these yeasts were amplified using the ITS1 and ITS4 primers as described above. The PCR products were purified using a PCR purification kit (Bioneer, Sought Korea) and sequenced by an automated DNA Sequencer (ABI Prism 3730 Genetic Analyzer; Applied Biosystems) using the ABI PRISM BigDye Terminator Cycle Sequencing Ready Reaction kit (Applied Biosystems). Sequences were subjected to basic local alignment search tool (BLAST) analysis, and the final species identification was performed based on the comparison of sequences with relevant sequences deposited in GenBank (http://www.ncbi.nlm.nih.gov/BLAST).
4. Results
During the study period, a total number of 930 yeast isolates were recovered from a variety of clinical samples (one sample from each patient). According to the colony color on CHROMagar Candida, 607 (65.3%) isolates were identified as C. albicans, 83 (8.9%) as C. tropicalis, and 15 (1.6%) as C. krusei while 225 (24.2%) remained doubtful or unknown species because of producing unspecified colony colors. As a result of PCR-RFLP identification, in addition to confirming the identified C. albicans and C. tropicalis strains by chromogenic medium, 142 (15.3%) were recognized as C. parapsilosis sensu lato, 52 (5.6%) as C. glabrata, and 15 (1.6%) as C. krusei based on the size of bands created by the Msp1 enzyme, and 31 (3.3%) remained unknown due to lacking the specified cutting sites of the enzyme. Finally, 30 isolates of the uncommon yeasts were identified to the species level by sequence analysis of ITS1-5.8SrDNA-ITS2 region and were recognized as Kluyveromyces marxianus (Candida kefyr) (n = 12, 38.7% of the 30 rare sequenced species), followed by (Clavispora (Candida) lusitaniae) (n = 8, 25.8%), C. intermedia (n = 3, 9.7%), C. orthopsilosis, Meyerozyma (Candida) guilliermondii and Trichosporon asahii (each n = 2 (6.5%), and Magnusiomyces capitatus (n = 1, 3.2%) (Table 1).
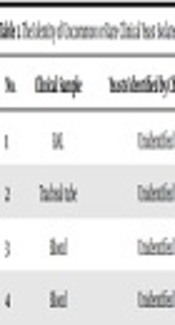
The Identity of Uncommon or Rare Clinical Yeast Isolates Determined by ITS-Sequencing
| No. | Clinical Sample | Yeasts Identified by Chromagar | Yeasts Identified by PCR-RFLP | Yeasts Identified by Sequencing |
|---|---|---|---|---|
| 1 | BAL | Unidentified | C. kefyr/C. famata | Kluyveromyces marxianus (C. kefyr) |
| 2 | Tracheal tube | Unidentified | C. kefyr/C. famata | Kluyveromyces marxianus (C. kefyr) |
| 3 | Blood | Unidentified | C. kefyr/C. famata | Kluyveromyces marxianus (C. kefyr |
| 4 | Blood | Unidentified | C. kefyr/C. famata | Kluyveromyces marxianus (C. kefyr) |
| 5 | Urine | Unidentified | C. kefyr/C. famata | Kluyveromyces marxianus (C. kefyr) |
| 6 | Throat | Unidentified | C. kefyr/C. famata | Kluyveromyces marxianus (C. kefyr) |
| 7 | Urine | Unidentified | C. kefyr/C. famata | Kluyveromyces marxianus (C. kefyr) |
| 8 | Blood | Unidentified | C. kefyr/C. famata | Kluyveromyces marxianus (C. kefyr) |
| 9 | Stool | Unidentified | C. kefyr/C. famata | Kluyveromyces marxianus (C. kefyr) |
| 10 | BAL | Unidentified | C. kefyr/C. famata | Kluyveromyces marxianus (C. kefyr) |
| 11 | Blood | Unidentified | C. kefyr/C. famata | Kluyveromyces marxianus (C. kefyr) |
| 12 | Stool | Unidentified | C. kefyr/C. famata | Kluyveromyces marxianus (C. kefyr) |
| 13 | Blood | Unidentified | C. lusitaniae/C. intermedia/C. rugosa | Clavispora lusitaniae |
| 14 | Urine | Unidentified | C. lusitaniae/C. intermedia/C. rugosa | Clavispora lusitaniae |
| 15 | Blood | Unidentified | C. lusitaniae/C. intermedia/C. rugosa | Clavispora lusitaniae |
| 16 | Blood | Unidentified | C. lusitaniae/C. intermedia/C. rugosa | Clavispora lusitaniae |
| 17 | Urine | Unidentified | C. lusitaniae/C. intermedia/C. rugose | Clavispora lusitaniae |
| 18 | Blood | Unidentified | C. lusitaniae/C. intermedia/C. rugosa | Clavispora lusitaniae |
| 19 | Blood | Unidentified | C. lusitaniae/C. intermedia/C. rugosa | Clavispora lusitaniae |
| 20 | Blood | Unidentified | C. lusitaniae/C. intermedia/C. rugosa | Clavispora lusitaniae |
| 21 | Blood | Unidentified | C. lusitaniae/C. intermedia/C. rugosa | Candida intermedia |
| 22 | Blood | Unidentified | C. lusitaniae/C. intermedia/C. rugosa | Candida intermedia |
| 23 | Blood | Unidentified | C. lusitaniae/C. intermedia/C. rugosa | Candida intermedia |
| 24 | Blood | Unidentified | C. parapsilosis | Candida orthopsilosis |
| 25 | Blood | Unidentified | C. parapsilosis | Candida orthopsilosis |
| 26 | Urine | Unidentified | Unknown | Meyerozyma guilliermondii |
| 27 | Nail | Unidentified | Unknown | Meyerozyma guilliermondii |
| 28 | Blood | Unidentified | Unknown | Meyerozyma guilliermondii |
| 29 | Blood | Unidentified | Unknown | Trichosporon asahii |
| 30 | Blood | Unidentified | Unknown | Magnusiomyces capitatus |
5. Discussion
Because of unspecified and variable clinical signs that occur late in the course of the disease, and due to the lack of sensitive and specific diagnostic methods, IFIs are often fatal (9). The major practice for the diagnosis of invasive fungal infections is based on conventional tests such as direct microscopy, histopathology, and culture. Advanced methods such as DNA detection and molecular-based methods should be done to heighten the ability of diagnosis of infections. In the present study, apart from five major common Candida species, the total frequency of the rare yeasts was 3.2%. This rate was about 9% in Qatar, another country in the Persian Gulf region (15). The overall frequency of uncommon Candida species is < 10% (16, 17). C. kefyr was the most frequent uncommon Candida species isolated in our study, followed by C. lusitaniae, C. intermedia, and C. orthopsilosis. In a large study on non-albicans Candida species carried out by Pfaller et al., the rank order of the most frequent species was C. glabrata > C. parapsilosis > C. tropicalis > C. krusei > C. lusitaniae > C. dubliniensis > C. guilliermondii (5). In Taiwan, C. guilliermondii, C. curvata, C. pelliculosa, and C. lusitaniae were the most uncommon Candida species (18). In addition, in a large study of uncommon Candida species in Texas, USA, C. guilliermondii, C. lusitaniae, C. kefyr, C. famata, and C. dubliniensis were found as the uncommon causes of candidemia (16). In the study of candidemia by uncommon Candida species in pediatrics in Iran, C. orthopsilosis, C. glabrata, C. lusitaniae, C. kefyr, C. dubliniensis, and C. intermedia were responsible for 12% of all cases of candidemia (4). These findings show that the distribution and frequency of uncommon Candida species are related to the geographic region and the population of patients. Due to the low incidence of infections caused by uncommon yeast species, the information on their clinical significance and microbial epidemiology is limited.
According to Table 1, uncommon or rare yeast species in this study were mostly isolated from blood and mucosal sites of patient bodies. Kluyveromyces marxianus (teleomorph of C. kefyr) is reported twice from patients in oncohematology wards, and most patients had myeloid or lymphoblastic leukemia. It is unknown why this species is an emerging fungal flora of neutropenic patients and causes outbreaks in hematology wards (19). C. lusitaniae causing about 1% of candidemia has been known as a notorious pathogen having initial resistance or rapid development of resistance to amphotericin B. It may present as breakthrough infection in immunocompromised patients on amphotericin B therapy (20). There is evidence that colony color switching on CHROMagar medium may occur during the course of therapy with amphotericin B; therefore, the presence of colony variants on the chromogenic medium should be investigated as a signal for the emergence of amphotericin B resistance in C. lusitaniae (20). Three cases of C. intermedia were reported in two separated studies in catheter-related candidemia (21, 22). In our study, C. intermedia was isolated from three blood samples of a three-year-old girl with two episodes of candidemia, who was implicated with cerebral palsy and respiratory disorders hospitalized in the PICU for about three months. Another case was related to a three-year-old girl with a gastrointestinal disorder that had undergone abdominal surgery. They survived after amphotericin B therapy. C. orthopsilosis seems to be less virulent than C. parapsilosissensu stricto and patients infected by C. orthopsilosis are more immunocompromised. These species are frequently related to bloodstream infections, particularly in pediatric patients (23). Overall, candidemia caused by C. parapsilosis sensu lato is often associated with the use of central venous catheters, parenteral nutrition, and the contamination of the hands of healthcare workers because of its high affinity to and capacity of being colonized on prosthetic materials and human skin (24, 25). Two cases of fungemia by C. orthopsilosis were found in this study. The patients were a 12-year-old girl and an 18-day-old neonate, hospitalized in the ICU for metabolic disease and prematurity, respectively, both of whom had central venous lines, parenteral nutrition, and intubation. Unfortunately, both patients died in spite of receiving amphotericin B therapy.
Meyerozyma guilliermondii (formerly Pichia guilliermondii) as the teleomorph C. guilliermondii was isolated from the urine sample of a premature neonate with congenital renal anomaly and another from nail scalping of a 62-year-old diabetic man. This species is a normal constituent of the human microbial flora and it is reported that 75% of C. guilliermondii isolates demonstrated reduced susceptibility to fluconazole (26). This species is increasingly reported in Latin America and a large pseudo-outbreak of C. guilliermondii fungemia was reported in Brazil (27).
Rare opportunistic non-Candida yeast isolates in our study were Trichosporon asahii and Magnusiomyces capitatus whilst in the study of Chitasombat et al., Rhodotorula, Trichosporon, Saccharomyces cerevisiae, Geotrichum, Pichia anomala, and Malassezia furfur were isolated (28). Trichosporon asahii was isolated from blood specimens of two patients; an infant with very low birth weight and prematurity who was bedridden in the NICU for two months and an 82-year-old diabetic woman with lower limb fracture that resulted in lower limb wound. In India, the majority of systemic trichosporonosis have been reported in diabetic patients with lower limb wounds (29). This species is the most frequently encountered species causing 84% of invasive Trichosporon infections (29). The importance of Trichosporon species are the inefficiency of amphotericin B and echinocandins therapy, particularly in the occurrence of breakthrough infections in patients receiving micafungin (30). Magnusiomyces capitatus (the anamorph Saprochaete capitis previously named Geotrichum capitatum, Trichosporon capitatum, or Blastoschizomyces capitatus) (9), as a colonizer of human skin and mucosa, was isolated from the BAL specimen of an 85-year-old man with chronic obstructive pulmonary disease hospitalized in the ICU. Despite the administration of amphotericin B and fluconazole, the patient died because of invasive infection due to immunodeficiency related to age and corticosteroid therapy. This species is an emerging opportunistic yeast in the Mediterranean region that causes a diverse spectrum of infections with high mortality rate regardless of antifungal treatment (31). However, between 1977 and 2013, around 104 cases of Saprochaete capitata and related species were reported in the English literature (32).
As the limitations of our work, it should be emphasized that yeast isolates were randomly collected in different periods from different healthcare centers; therefore, they are not the exact representation of the distribution of yeast species and clinical forms of candidiasis. In addition, the relationships between the yeast species and the clinical status of the disease and outcome were not investigated. The determination of antifungal susceptibility profile of the uncommon isolated yeast was not tested in this study.
5.1. Conclusions
The isolation of emerging or less common yeast species, which cause a variety of infections from superficial to systemic infections, is increasingly reported. Since these uncommon yeast species may exhibit low susceptibility to some antifungal agents, the use of reliable methods for accurate identification and subsequently correct management is necessary.
Acknowledgements
References
-
1.
Pfaller MA, Diekema DJ. Epidemiology of invasive candidiasis: A persistent public health problem. Clin Microbiol Rev. 2007;20(1):133-63. [PubMed ID: 17223626]. [PubMed Central ID: PMC1797637]. https://doi.org/10.1128/CMR.00029-06.
-
2.
Sardi JC, Scorzoni L, Bernardi T, Fusco-Almeida AM, Mendes Giannini MJ. Candida species: Current epidemiology, pathogenicity, biofilm formation, natural antifungal products and new therapeutic options. J Med Microbiol. 2013;62(Pt 1):10-24. [PubMed ID: 23180477]. https://doi.org/10.1099/jmm.0.045054-0.
-
3.
Charsizadeh A, Nikmanesh B, Ahmadi B, Jalalizand N, Jafari Z, Rahmani M, et al. Frequency of candida species isolated from patients in Children’s Medical Center, Tehran, Iran. Prevalence. 2017;27:9.
-
4.
Charsizadeh A, Mirhendi H, Nikmanesh B, Eshaghi H, Rahmani M, Farhang A, et al. Candidemia in children caused by uncommon species of Candida. Arch Pediatr Infect Dis. 2018;6(2). https://doi.org/10.5812/pedinfect.11895.
-
5.
Pfaller MA, Andes DR, Diekema DJ, Horn DL, Reboli AC, Rotstein C, et al. Epidemiology and outcomes of invasive candidiasis due to non-albicans species of Candida in 2,496 patients: Data from the prospective antifungal therapy (PATH) registry 2004-2008. PLoS One. 2014;9(7). e101510. [PubMed ID: 24991967]. [PubMed Central ID: PMC4081561]. https://doi.org/10.1371/journal.pone.0101510.
-
6.
Skiada A, Pavleas I, Drogari-Apiranthitou M. Rare fungal infectious agents: A lurking enemy. F1000Res. 2017;6:1917. [PubMed ID: 29152230]. [PubMed Central ID: PMC5664977]. https://doi.org/10.12688/f1000research.11124.1.
-
7.
Charsizadeh A, Mirhendi H, Nikmanesh B, Eshaghi H, Makimura K. Microbial epidemiology of candidaemia in neonatal and paediatric intensive care units at the Children's Medical Center, Tehran. Mycoses. 2018;61(1):22-9. [PubMed ID: 28872714]. https://doi.org/10.1111/myc.12698.
-
8.
Vaezi A, Fakhim H, Khodavaisy S, Alizadeh A, Nazeri M, Soleimani A, et al. Epidemiological and mycological characteristics of candidemia in Iran: A systematic review and meta-analysis. J Mycol Med. 2017;27(2):146-52. [PubMed ID: 28318900]. https://doi.org/10.1016/j.mycmed.2017.02.007.
-
9.
Arendrup MC, Boekhout T, Akova M, Meis JF, Cornely OA, Lortholary O, et al. ESCMID and ECMM joint clinical guidelines for the diagnosis and management of rare invasive yeast infections. Clin Microbiol Infect. 2014;20 Suppl 3:76-98. [PubMed ID: 24102785]. https://doi.org/10.1111/1469-0691.12360.
-
10.
Miceli MH, Diaz JA, Lee SA. Emerging opportunistic yeast infections. Lancet Infect Dis. 2011;11(2):142-51. [PubMed ID: 21272794]. https://doi.org/10.1016/S1473-3099(10)70218-8.
-
11.
Vazquez JA. Rhodotorula, malassezia, trichosporon, and other yeast-like fungi. In: Dismukes WE, Pappas PG, Sobel JD, editors. Clinical Mycology. Oxford University Press; 2003. 206 p.
-
12.
White TJ, Bruns T, Lee SA, Taylor JL. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, White TJ, editors. PCR protocols: A guide to methods and application. Elsevier; 1989. p. 315-22. https://doi.org/10.1016/B978-0-12-372180-8.50042-1.
-
13.
Millar BC, Jiru X, Moore JE, Earle JA. A simple and sensitive method to extract bacterial, yeast and fungal DNA from blood culture material. J Microbiol Methods. 2000;42(2):139-47. [PubMed ID: 11018270]. https://doi.org/10.1016/S0167-7012(00)00174-3.
-
14.
Mohammadi R, Mirhendi H, Rezaei-Matehkolaei A, Ghahri M, Shidfar MR, Jalalizand N, et al. Molecular identification and distribution profile of Candida species isolated from Iranian patients. Med Mycol. 2013;51(6):657-63. [PubMed ID: 23470036]. https://doi.org/10.3109/13693786.2013.770603.
-
15.
Taj-Aldeen SJ, AbdulWahab A, Kolecka A, Deshmukh A, Meis JF, Boekhout T. Uncommon opportunistic yeast bloodstream infections from Qatar. Med Mycol. 2014;52(5):552-6. [PubMed ID: 24934803]. https://doi.org/10.1093/mmycol/myu016.
-
16.
Jung DS, Farmakiotis D, Jiang Y, Tarrand JJ, Kontoyiannis DP. Uncommon Candida species fungemia among cancer patients, Houston, Texas, USA. Emerg Infect Dis. 2015;21(11):1942-50. [PubMed ID: 26488845]. [PubMed Central ID: PMC4625381]. https://doi.org/10.3201/eid2111.150404.
-
17.
Pfaller M, Neofytos D, Diekema D, Azie N, Meier-Kriesche HU, Quan SP, et al. Epidemiology and outcomes of candidemia in 3648 patients: Data from the prospective antifungal therapy (PATH Alliance(R)) registry, 2004-2008. Diagn Microbiol Infect Dis. 2012;74(4):323-31. [PubMed ID: 23102556]. https://doi.org/10.1016/j.diagmicrobio.2012.10.003.
-
18.
Liu WL, Lai CC, Li MC, Wu CJ, Ko WC, Hung YL, et al. Clinical manifestations of candidemia caused by uncommon Candida species and antifungal susceptibility of the isolates in a regional hospital in Taiwan, 2007-2014. J Microbiol Immunol Infect. 2017. [PubMed ID: 28886952]. https://doi.org/10.1016/j.jmii.2017.08.007.
-
19.
Sendid B, Lacroix C, Bougnoux ME. Is Candida kefyr an emerging pathogen in patients with oncohematological diseases? Clin Infect Dis. 2006;43(5):666-7. [PubMed ID: 16886166]. https://doi.org/10.1086/506573.
-
20.
McClenny NB, Fei H, Baron EJ, Gales AC, Houston A, Hollis RJ, et al. Change in colony morphology of Candida lusitaniae in association with development of amphotericin B resistance. Antimicrob Agents Chemother. 2002;46(5):1325-8. [PubMed ID: 11959563]. [PubMed Central ID: PMC127144]. https://doi.org/10.1128/AAC.46.5.1325-1328.2002.
-
21.
Ruan SY, Chien JY, Hou YC, Hsueh PR. Catheter-related fungemia caused by Candida intermedia. Int J Infect Dis. 2010;14(2):e147-9. [PubMed ID: 19497773]. https://doi.org/10.1016/j.ijid.2009.03.015.
-
22.
Hasejima N, Kamei K, Matsubayashi M, Kawabe R, Shimura C, Hijikata N, et al. The first case of bloodstream infection by Candida intermedia in Japan: The importance of molecular identification. J Infect Chemother. 2011;17(4):555-8. [PubMed ID: 21302127]. https://doi.org/10.1007/s10156-011-0215-4.
-
23.
Ruiz LS, Khouri S, Hahn RC, da Silva EG, de Oliveira VK, Gandra RF, et al. Candidemia by species of the Candida parapsilosis complex in children's hospital: Prevalence, biofilm production and antifungal susceptibility. Mycopathologia. 2013;175(3-4):231-9. [PubMed ID: 23404576]. https://doi.org/10.1007/s11046-013-9616-5.
-
24.
Ataides FS, Costa CR, Souza LK, Fernandes O, Jesuino RS, Silva Mdo R. Molecular identification and antifungal susceptibility profiles of Candida parapsilosis complex species isolated from culture collection of clinical samples. Rev Soc Bras Med Trop. 2015;48(4):454-9. [PubMed ID: 26312937]. https://doi.org/10.1590/0037-8682-0120-2015.
-
25.
Constante CC, Monteiro AA, Alves SH, Carneiro LC, Machado MM, Severo LC, et al. Different risk factors for candidemia occur for Candida species belonging to the C. parapsilosis complex. Med Mycol. 2014;52(4):403-6. [PubMed ID: 24782105]. https://doi.org/10.1093/mmy/myt034.
-
26.
Savini V, Catavitello C, Onofrillo D, Masciarelli G, Astolfi D, Balbinot A, et al. What do we know about Candida guilliermondii? A voyage throughout past and current literature about this emerging yeast. Mycoses. 2011;54(5):434-41. [PubMed ID: 21039941]. https://doi.org/10.1111/j.1439-0507.2010.01960.x.
-
27.
Nucci M, Queiroz-Telles F, Tobon AM, Restrepo A, Colombo AL. Epidemiology of opportunistic fungal infections in Latin America. Clin Infect Dis. 2010;51(5):561-70. [PubMed ID: 20658942]. https://doi.org/10.1086/655683.
-
28.
Chitasombat MN, Kofteridis DP, Jiang Y, Tarrand J, Lewis RE, Kontoyiannis DP. Rare opportunistic (non-Candida, non-Cryptococcus) yeast bloodstream infections in patients with cancer. J Infect. 2012;64(1):68-75. [PubMed ID: 22101079]. [PubMed Central ID: PMC3855381]. https://doi.org/10.1016/j.jinf.2011.11.002.
-
29.
Slavin MA, Chakrabarti A. Opportunistic fungal infections in the Asia-Pacific region. Med Mycol. 2012;50(1):18-25. [PubMed ID: 21905945]. https://doi.org/10.3109/13693786.2011.602989.
-
30.
Liao Y, Lu X, Yang S, Luo Y, Chen Q, Yang R. Epidemiology and outcome of trichosporon fungemia: A review of 185 reported cases from 1975 to 2014. Open Forum Infect Dis. 2015;2(4):ofv141. [PubMed ID: 26566536]. [PubMed Central ID: PMC4630454]. https://doi.org/10.1093/ofid/ofv141.
-
31.
Subramanya Supram H, Gokhale S, Chakrabarti A, Rudramurthy SM, Gupta S, Honnavar P. Emergence of Magnusiomyces capitatus infections in Western Nepal. Med Mycol. 2016;54(2):103-10. [PubMed ID: 26483432]. https://doi.org/10.1093/mmy/myv075.
-
32.
Mazzocato S, Marchionni E, Fothergill AW, Sutton DA, Staffolani S, Gesuita R, et al. Epidemiology and outcome of systemic infections due to saprochaete capitata: Case report and review of the literature. Infection. 2015;43(2):211-5. [PubMed ID: 25078793]. https://doi.org/10.1007/s15010-014-0668-3.