1. Background
Staphylococcus aureus (S. aureus) as a Gram-positive pathogen is known as the main cause of infections ranging from minor skin infections to serious infections including osteomyelitis, bacteremia, infective endocarditis, necrotizing pneumonia, and toxic shock syndrome (TSS) (1). There are several virulence factors in S. aureus including cell surface virulence factors (capsular polysaccharides), exoproteins (cytolysins), and superantigens (TSST-1) (2). Superantigens are responsible for a number of human diseases such as food poisoning and TSS. These bacterial toxins can activate T cell receptor (TCR), immunoreceptors, major histocompatibility complex (MHC) class II, and release cytokines (3).
TSS is a polysystemic disease with symptoms including hypotension, fever, shock, skin rash, and skin peeling. The cause of TSS in most individuals with wound infection is the MRSA colonization (4). Based on recent reports, there is a direct relationship between the use of tampon by women and the incidence of TSS in the healthy population (3). Conventionally, S. aureus colonization occurs in different parts of the body such as perineal, axillary, and nasal regions called the nasal carriage of S. aureus (5).
The colonization of S. aureus is not often the main reason for infection; however, it is a known risk factor for the emergence of subsequent infections so that it can act as a reservoir of clinical infections in colonized individuals (6, 7).
At present, the appearance of antibiotic-resistant S. aureus strains called methicillin-resistant S. aureus (MRSA) has been known as a serious threat in hospital and community settings. MRSA infections are mostly associated with high morbidity and mortality and the increased length of hospital stay, particularly in individuals with HIV, cancer, rheumatoid arthritis, and diabetes (8).
The mecA gene in Staphylococcus species is responsible for resistance to methicillin. It is located on chromosomal cassette mec and encodes a low-affinity penicillin-binding protein (PBP2a). Therefore, the MRSA strains are resistant to all beta-lactam antibiotics (9, 10). In recent years, novel MRSA strains have emerged as oxacillin-susceptible mecA-positive S. aureus (OS-MRSA) strains that carry the mecA gene but are sensitive to oxacillin; thus, they could be wrongly identified as MSSA (11). At present, the spread of the OS-MRSA isolates is considered to be due to their misidentification as MSSA and treatment failure with β-lactam antibiotics (12).
2. Objectives
This study aimed to isolate OS-MRSA and identify the Tsst-1 virulence factor among high school students.
3. Methods
3.1. Bacteria Identification
The nasal swab samples were obtained from 400 randomly selected healthy students (200 boys and 200 girls) from six different high schools in Tabriz city from January 1, 2018, to March 1, 2018. The obtained samples were cultured in selective media (blood agar and mannitol salt agar) and incubated overnight at 37ºC. S. aureus strains were identified by conventional microbiological and biochemical methods (colony morphology, Gram staining, catalase test, and coagulase test) (13).
3.2. Antimicrobial Susceptibility Testing
The susceptibility pattern of the isolates was determined using 12 antibiotics based on the Kirby Bauer disk diffusion method following the Clinical Laboratory Standards Institute (CLSI) guidelines (10). The methicillin sensitivity test was carried out based on resistance to cefoxitin (30 μg/disc). The used antibiotic disks were as follows: erythromycin (15 μg), chloramphenicol (30 μg), oxacillin (1 μg), ciprofloxacin (5 μg), clindamycin (2 μg), penicillin (6 μg), amoxicillin/clavulanic acid (20/10 μg), cefazolin (30 μg), vancomycin (30 μg), cefoxitin (30 μg), ampicillin (10 μg), and novobiocin (5 μg). To perform the disk diffusion test, bacterial concentrations equal to 0.5 McFarland were prepared to inoculate Muller-Hinton agar plats. The antibiotic disks were incubated with the inoculated plates at 37ºC overnight. All antibiotics were produced by Biomaxima (Poland).
3.3. Primer Designing
The specific primers of TSST-1 and mecA genes were designed using Gene Runner software based on the S. aureus genome sequences available in the Gene Bank database. The specificity of the designed primers was evaluated using Primer-BLAST.
3.4. DNA Extraction and PCR Amplification
DNA was extracted from individual colonies using the boiling method. In this method, pure colonies were suspended in 300 μL of TE buffer (10 mM Tris, 1 mM EDTA); then, cell suspensions were boiled at 95ºC and centrifuged. The quantification of the extracted DNA was carried out via spectroscopy at 260 nm. The DNA purity was determined by the ratio of absorbance at 260/280. Then, the PCR was carried out to examine the presence of mecA and TSST-1 genes using specific primers (Table 1). The PCR products were analyzed by the agarose gel electrophoresis. For further corroboration, the PCR product of each gene was analyzed by sequencing.
| Primer | Primer Sequence (5’→3’) | Temperature, ºC | Cycle No. | Size, bp |
|---|---|---|---|---|
| mecA | F:5’-AGAAATGACTGAACGTCC-3’ | 48 | 35 | 305 |
| R:5’-ATTCCACATTGTTTCGGTC-3’ | ||||
| TSST-1 | F: 5’-ACAAGCGCTATTTTTATTTCAG-3’ | 49 | 30 | 271 |
| R: 5’-CCCATCCCCAACCACTTTT-3’ |
4. Results
4.1. Bacterial Identification
In total, 400 nasal samples were obtained from male and female students of six different high schools in Tabriz. Based on the results, 60 (15%) students were positive for S. aureus; 9.5% (38/400) of the S. aureus-positive isolates belonged to male students and 5.5% (22/400) to female students. The statistical analysis showed that the frequency of S. aureus was dependent on gender (P < 0.05).
4.2. Antimicrobial Susceptibility
The antibiotic resistance patterns were determined using 12 antibiotics by the disc diffusion method. All S. aureus isolates were resistant to ampicillin and 98.33% to penicillin. All of the 60 isolates were sensitive to vancomycin. Based on the results, 18.34% (11/60) of the isolates were resistant to cefoxitin (MRSA) and 11.67% (7/60) were resistant to oxacillin. The results are summarized in Table 2. From 60 S. aureus-positive strains, 19 (31.66%) showed multi-drug resistant (MDR) property. A significant correlation was observed between the presence of the mecA gene and resistance to ampicillin, penicillin, oxacillin, ciprofloxacin, erythromycin, cefoxitin, and amoxicillin/clavulanic acid (P < 0.05).
| Antibiotics | Resistant, % | Intermediate, % | Sensitive, % |
|---|---|---|---|
| Erythromycin | 18.34 | 10 | 71.66 |
| Chloramphenicol | 3.33 | 3.33 | 93.34 |
| Oxacillin | 11.67 | 1.66 | 86.67 |
| Ciprofloxacin | 5 | 26.67 | 68.33 |
| Clindamycin | 6.67 | 3.33 | 90 |
| Ampicillin | 100 | - | - |
| Amoxicillin/clavulanic acid | 36.67 | - | 63.33 |
| Penicillin | 98.33 | - | 1.67 |
| Cefoxitin | 18.34 | - | 81.66 |
| Cefazolin | 5 | 1.67 | 93.33 |
| Vancomycin | - | - | 100 |
4.3. Detection of mecA and TSST-1 Genes by PCR
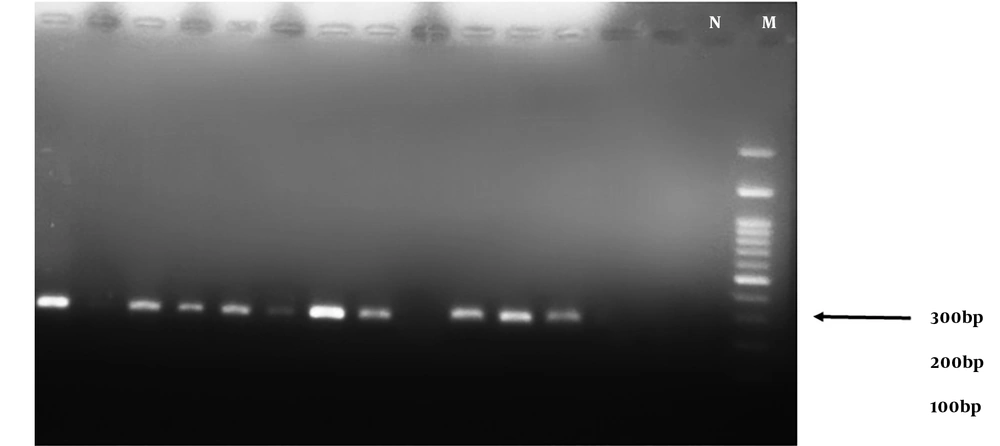
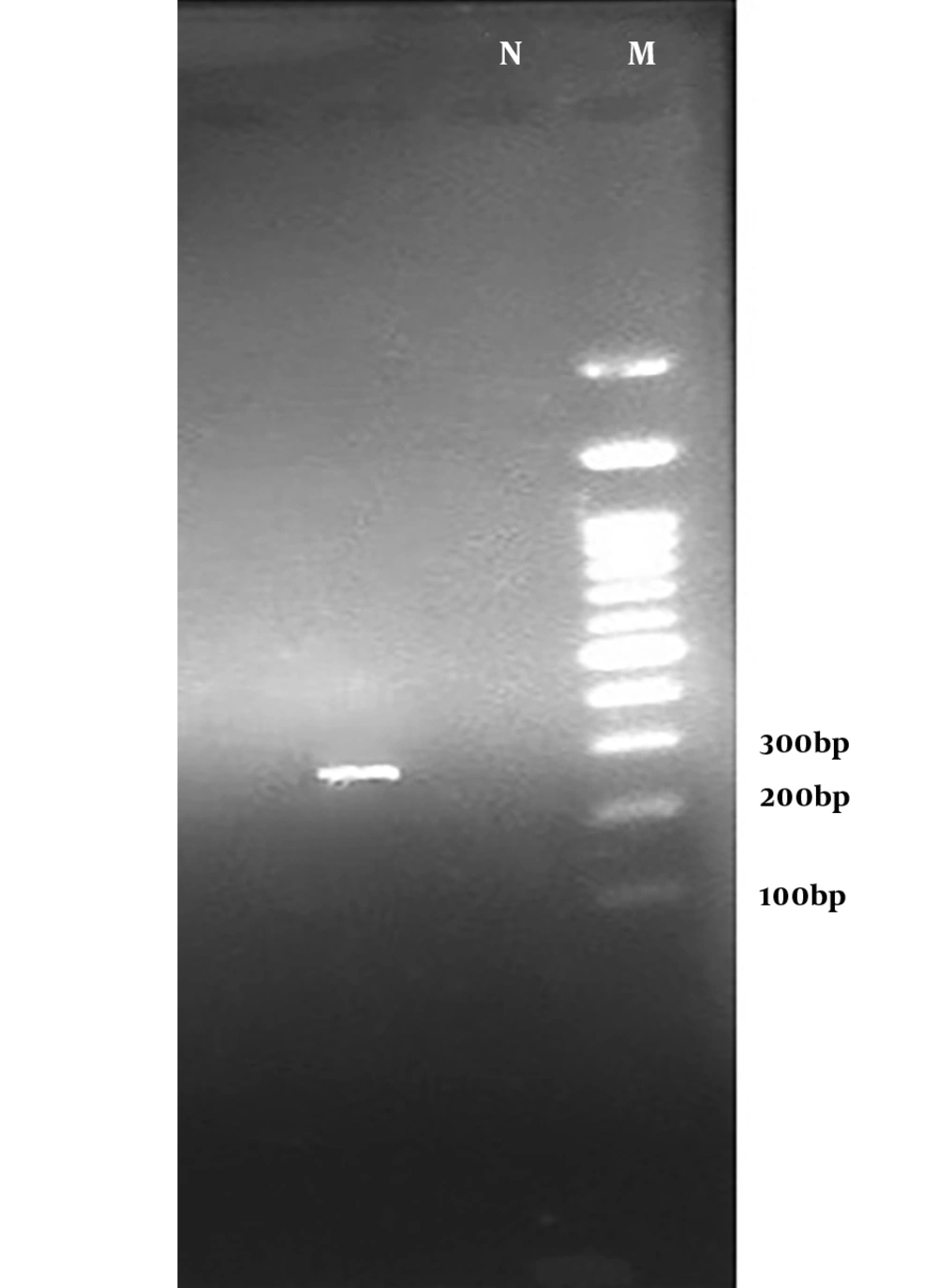
The results of the PCR assay performed on S. aureus isolates showed that 54.54% (36/60) of the strains were positive for the mecA gene (Figure 1) while only 11.67% were resistant to oxacillin. These results indicate the emergence of OS-MRSA for the first time in the healthy population of Iran. Based on the results of PCR, the TSST-1 gene was detected as an expected band of 271 bp (Figure 2) only in 1.5% (one sample) of the strains from an MRSA isolate belonging to a girl student.
4.4. Sequencing Analysis
Since the TSST-1-positive strain was the first report in the study region, the sequence was amplified by TSST-1 specific primers, subjected to sequencing, and analyzed by NCBI BLAST. The results showed that the TSST-1-positive isolate had 100% homology to the TSST-1 gene previously reported in an S. aureus strain (Sequence ID: AB084255.1).
5. Discussion
In the last two decades, the epidemiology of MRSA infections has changed from healthcare-associated (HA)-MRSA to community-associated (CA)-MRSA as a causative agent of infection in clinical settings (14-16). On the other hand, the prevalence of MRSA infections has emerged as a serious problem since MRSA strains are resistant to most antibiotics, particularly β-lactams. Based on recent studies, the nasal colonization of S. aureus plays a major role in the pathogenesis and epidemiology of infections (17). At present, the prevalence of the MRSA nasal carriage is increasing in the healthy population. For instance, in a study performed in Jordan, the frequency of the MRSA nasal carriage was 7.5% in a healthy population (18). In a similar study conducted in Western Nepal, 32 out of 204 (15.7 %) healthcare workers were S. aureus carriers, of which 21.9% (7 cases) were MRSA (19). In the present study, the frequency of nasal carriage of S. aureus was examined in a healthy population. Based on the results, 60 out of 400 (15%) students were nasal carriers of S. aureus and 2.18% (11/400) of the students were MRSA-positive based on the phenotypic test.
Our results are consistent with previous results reported by Khanal et al. in 2015 that highlighted the MRSA nasal carriage rate of 3.4% (7/204) among health care workers in Western Nepal (19). In the present study, 18.34% of the S. aureus-positive samples were resistant to erythromycin and cefoxitin and 11.67% were resistant to oxacillin. These findings are not consistent with a previous study of nasal carriers of S. aureus in nursing students that reported the rate of S. aureus positivity as 32.5%, MRSA as 10%, and resistance to erythromycin as 30% (20). According to our results, 6.67% of the isolates were resistant to clindamycin that contradicts the findings of da Silva et al. in Pernambuco; they assessed resistance to clindamycin among nursing staff in a teaching hospital and reported that 11.9% of the nasal carriers were resistant to clindamycin (21). Currently, a notable increase is reported in the prevalence of MDR among MRSA isolates that imposes a serious menace on public health (22).
In the present study, from 60 S. aureus-positive isolates, 31.66% (19 cases) were MDR-positive. This result is consistent with a previous study in Kashan, Iran, that reported 26.3% of healthy children were nasal carriers of S. aureus, 29.3% were MDR-positive, and 35.9% were MRSA-positive (23).
The results of PCR indicated that the mecA gene was present in 54.54% (36/60) of the S. aureus isolates while only 11.67% of the S. aureus isolates were resistant to oxacillin. As a result, 6.25% of the students were OS-MRSA carriers. These results indicate the increasing prevalence of OS-MRSAs in the healthy population of Iran concerning the results of a study performed in 2013 on 173 nurses in Iran that reported 4.6% of the cases were MRSA carriers and only 1.15% of the MRSA isolates were OS-MRSA (24). In a study performed on healthcare workers in two African countries, 2.4% of the individuals were OS-MRSA carriers that can transfer it to patients as the reservoirs of OS-MRSA (25). Recent studies indicate an increase in the prevalence of oxacillin-susceptible mecA-positive S. aureus in human infections (26, 27).
Since OS-MRSA isolates carry the mecA gene but are sensitive to oxacillin, they could be wrongly identified as MSSA if the detection of the mecA gene is not tried (28). In the present study, one of the isolates (1.5%), which was an MRSA-MDR strain, was positive for the TSST-1 gene. This result is consistent with the results by Hogan et al. in 2016 regarding the detection of the TSST-1 gene among students in Madagascar in which, 1.9% of the students were positive for the TSST-1 gene (29).
5.1. Conclusions
Our findings highlight a high prevalence of OS-MRSA in high school students of Tabriz, Northwest of Iran. Nevertheless, the combination of genetic and phenotypic tests could be essential to accurately detect MRSA.
5.2. Limitations
This study did not examine the molecular characteristics of OS-MRSA and TSST-1-positive MRSA strains.