Abstract
Background:
Bacterial resistance is a worldwide phenomenon that can disrupt the treatment of many different infectious diseases. Identifying drug-resistant bacteria is very important in different aspects, such as choosing the appropriate antibiotics, accelerating treatment, reducing the costs of treatment, and preventing antibiotic resistance.Objectives:
Therefore, the present study was aimed to investigate the prevalence of drug-resistant Gram-negative bacteria in a teaching hospital in Tehran, Iran.Methods:
In the present cross-sectional study, all clinical specimens that were obtained from patients admitted to the Imam Hossein Hospital for infections caused by Gram-negative bacteria during 2017 were included. The pattern of antibiotic resistance was determined by the disk diffusion test as recommended by the Clinical Laboratory and Standards Institute (CLSI) guideline.Results:
The result of the culture for 295 patients under study was reported as positive for Gram-negative bacteria. The most frequent Gram-negative bacteria were Escherichia coli (31.2%), followed by Klebsiella spp. (20.3%) and Pseudomonas spp. (13.2). The most antibiotic resistance was observed against cephalexin, ceftriaxone, and cefotaxime.Conclusions:
Resistance in Gram-negative bacteria was relatively high in the current study. Establishment of better infection control policies and education of hospital staff, especially in the ICU are recommended for the prevention and control of drug-resistant pathogens in the health care settings.Keywords
1. Background
Widespread use of broad-spectrum antibiotics in recent years has led to the emergence of drug-resistant bacteria (1, 2). The spread of these bacteria poses a substantial threat to healthcare settings worldwide. Due to its public health implications, drug-resistant bacteria have been recently highlighted by the World Health Organization (WHO) as a global priority for investment in research and development of effective drugs (3). These microorganisms are even showing rising rates of resistance to new antibiotics, subsequently disrupting the treatment of many infectious diseases (4-6). Critical-priority drug-resistant Gram-negative bacteria included Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacteriaceae (3). Each of the aforementioned isolates has a different sensitivity to various antibiotics (5). For instance, in a study conducted in the US, Escherichia coli was mostly resistant to penicillin and cephalosporin (7). The observed resistance can be due to the production of beta-lactamase enzymes, decreasing permeability, and increasing active efflux of the antibiotic or mutation (7-11).
Identifying antibiotic-resistant bacteria is important in many different aspects such as choosing the appropriate antibiotics, accelerating the treatment, reducing the costs of treatment and also preventing antibiotic resistance.
2. Objectives
Thus, the present study was aimed to investigate the prevalence of drug-resistant Gram-negative bacteria in a teaching hospital in Tehran, Iran.
3. Methods
3.1. Samples and Setting
The present study was an observational-analytical study that was performed in a cross-sectional design on all the clinical samples of patients admitted to the Imam Hossein Hospital for infections caused by Gram-negative bacteria during 2017. Clinical samples include blood, sputum, cerebrospinal fluid, urine, chip discharge, pleural fluid, ascites, ocular, ulcer, and catheter fluid.
3.2. Bacterial Isolation
All specimens were inoculated on proper culture media (Hi-Media, India) within two hours. Only one isolate per patient was included. Methods used for identification of Gram-negative bacteria include an examination of colonial morphology, hemolytic characteristics on blood agar media, Gram stain, and biochemical tests (i.e. catalase, oxidase, coagulase, bile solubility, and spot indole).
3.3. Drug Susceptibility Testing
The isolated bacteria were inoculated on Mueller Hinton agar (Mast group Ltd, Merseyside, UK) and antimicrobial susceptibility testing was performed using the disk diffusion method as recommended by Clinical and Laboratory Standard Institute (CLSI No: M2-A9). Quality control was assured by concurrent testing with American Type Culture Collection (ATCC) strains including Eshershia coli ATCC 25922, Pseudomonas aeruginosa ATCC 27853 and Staphylococcus aureus ATCC 25923. The following antibiotics; amikacin (10 microgram), gentamicin (10 microgram), ciprofloxacin (5 microgram), levofloxacin (5 microgram), ceftriaxone (30 microgram), ceftazidime (30 microgram), cefepime (30 microgram), cefotaxime (30 microgram), imipenem (10 microgram), meropenem (10 microgram), and colistin sulfate (10 microgram) were used for drug susceptibility testing. The antibiotic disks were provided from Hi-Media group Ltd.
3.4. Statistical Analysis
All data were analyzed by the SPSS software (version 22, SPSS Inc., Chicago, IL, USA). Independent sample t-test was used to compare quantitative variables and chi-square was used for qualitative variables. All the statistical test results of less than 0.05 were considered as significant.
4. Results
In this study, the culture from 295 patients was reported as positive for Gram-negative pathogens. The participants of the study consisted of 144 men (48.8%) and 151 women (51.2%); the average age was 35.57 ± 9.91 with the age range of 15 to 60. The most frequent Gram-negative bacteria was E. coli with a frequency of 92 cases (31.2%), followed by Klebsiella spp. with the frequency of 60 cases (20.3%), and Pseudomonas spp. with the frequency of 39 cases (13.2%). The samples used in this study were taken from different fluids such as urine, feces, blood, sputum, wounds, catheters, cerebrospinal fluid, ascites, throat, tracheal and eye discharges, and pleural effusions; their frequency distribution are provided in Table 1.
| Culture Place | Values |
|---|---|
| Urinary | 101 (34.6) |
| Blood | 70 (24) |
| Sputum | 41 (14) |
| Wound | 28 (9.6) |
| catheter | 15 (5.1) |
| Trachea | 14 (4.8) |
| Cerebrospinal fluid | 7 (2.4) |
| Ascites | 5 (1.7) |
| Throat | 3 (1) |
| Eye | 2 (0.7) |
| Undetermined | 2 (0.7) |
| Pleura | 1 (0.3) |
| Stool | 1 (0.3) |
According to the drug susceptibility results, a high rate of resistance to cephalexin (100%), ceftriaxone (90%), and cefotaxime (73.0%) was observed. Likewise, resistance to imipenem amikacin and colistin was found to be 36.2%, 32.3%, and 7.5%, respectively. The frequency distribution of drug resistance in Gram-negative bacteria is shown in Table 2.
| Causes | Sensitive | Intermediate | Resistance | Total |
|---|---|---|---|---|
| Cephalexin | - | - | 105 (100) | 105 |
| Ceftriaxone | 2 (10) | - | 18 (90) | 20 |
| Cefotaxime | 49 (26.92) | - | 133 (73.08) | 182 |
| Ceftazidime | 62 (18.86) | - | 153 (71.16) | 215 |
| Cefepime | 54 (27.14) | 5 (2.51) | 140 (73.35) | 199 |
| Levofloxacin | 93 (34.31) | 5 (1.85) | 173 (63.84) | 271 |
| Ciprofloxacin | 91 (31.27) | 13 (7.56) | 178 (61.17) | 291 |
| Tobramycin | 2 (50) | - | 2 (50) | 4 |
| Meropenem | 127 (54.27) | - | 107 (45.73) | 234 |
| Gentamycin | 9 (24.57) | - | 7 (43.75) | 16 |
| Imipenem | 173 (61.56) | 6 (2.14) | 102 (36.30) | 281 |
| Amikacin | 164 (62.12) | 15 (5.68) | 85 (32.20) | 264 |
| Colistin | 110 (43.3) | - | 9 (7.56) | 119 |
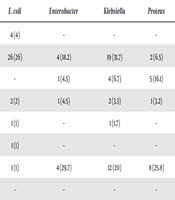
The distribution of Gram-negative bacteria based on the site of infection is shown in Table 3. E. coli was frequently isolated in urine samples (54%); Klebsiella spp. was frequently seen in the blood samples (31.7%), and Pseudomonas spp. was frequently seen in blood and tracheal samples (29.7).
| Acinobacter | Citrobacter | E. coli | Enterobacter | Klebsiella | Proteus | Pseudomonas | Serratia | Shigella | |
|---|---|---|---|---|---|---|---|---|---|
| Ascites | - | - | 4 (4) | - | - | - | 1 (2.6) | - | - |
| Blood | 11 (29.7) | - | 26 (26) | 4 (18.2) | 19 (31.7) | 2 (6.5) | 8 (29.7) | - | - |
| Catheter | 2 (5.4) | - | - | 1 (4.5) | 4 (6.7) | 5 (16.1) | 3 (7.7) | - | - |
| Cerebrospinal fluid | 1 (2.7) | - | 2 (2) | 1 (4.5) | 2 (3.3) | 1 (3.2) | - | - | - |
| Eye | - | - | 1 (1) | - | 1 (1.7) | - | - | - | - |
| Pleura | - | - | 1 (1) | - | - | - | - | - | - |
| Sputum | 9 (24.3) | - | 1 (1) | 4 (29.7) | 12 (20) | 8 (25.8) | 7 (17.9) | - | - |
| Stool | - | - | - | - | - | - | - | - | 1 (100) |
| Throat | 2 (5.4) | - | - | - | - | - | 1 (29.7) | - | - |
| Trachea Culture | 6 (16.2) | - | - | 1 (4.5) | 3 (5) | 2 (6.5) | 2 (29.7) | - | - |
| Undetermined | - | - | - | - | 2 (3.2) | - | - | - | - |
| Urinary | 3 (8.1) | 1 (100) | 54 (54) | 5 (22.7) | 14 (23.3) | 10 (32.3) | 13 (5.1) | 1 (100) | - |
| Wound | 3 (8.1) | - | 11 (11) | 6 (27.3) | 3 (5) | 3 (9.7) | 2 (5.1) | 1 (100) | 1 (100) |
| Total | 37 (100) | 1 (100) | 100 (100) | 22 (100) | 60 (100) | 31 (100) | 39 (100) |
The pattern of drug resistance of isolated Gram-negative bacteria is shown in Table 4.
| Ceftazidime | Ceftriaxone | Cefotaxime | Cefepime | Levofloxacin | Meropenem | Imipenem | Colistin | Amikacin | |
|---|---|---|---|---|---|---|---|---|---|
| Acinobacter | 22 (14.5) | - | 18 (13.6) | 18 (13.6) | 31 (18) | 32 (9.9) | 33 (32.4) | - | - |
| Citrobacter | - | - | - | - | 1 (6) | - | - | - | - |
| E. coli | 66 (43.4) | 6 (33.3) | 61 (47) | 61 (47) | 48 (27.9) | 3 (2.8) | 5 (4.9) | - | 12 (85.7) |
| Enterobacter | 10 (6.6) | 1 (6.5) | 10 (7.6) | 10 (7.6) | - | - | - | - | 1 (7.1) |
| Klebsiella | 27 (17.8) | 3 | 26 (12.19) | 26 (19.12) | 44 (25.6) | 36 (33.6) | 35 (34.3) | 9 (100) | - |
| Proteus | 18 (11.8) | 6 | 15 (11.4) | 15 (11.4) | 21 (12.2) | 13 (12.1) | 13 (12.7) | - | - |
| Pseudomonas | 8 (5.3) | 1 | 1(8) | 1 (8) | 27 (15.7) | 33 (21.5) | 16 (15.7) | - | - |
| Shigella | - | 1 | - | - | - | - | - | - | - |
| Serratia | - | - | - | - | - | - | - | - | - |
5. Discussion
In this study, the most frequent Gram-negative bacteria were E. coli followed by Klebsiella spp and Pseudomonas spp. In the similar studies conducted in Arak (12), Bandar Abbas (13), and Khorramabad (14), the most prevalent strains obtained were E. coli, which are similar to the results of the present study. However, in the study performed in Besat Hospital the most prevalent strain was Klebsiella spp. It seems that the observed discrepancy in the results of the Besat Hospital study was due to the fact that their samples were obtained from patients admitted to the ICU (15). Regarding the high prevalence of E. coli, it can be concluded that despite the numerous antibiotic treatments, this bacteria is still the most prevalent Gram-negative bacilli. Regarding the high prevalence of this isolate in the urine samples, control, treatment, and prevention of urinary tract infections is highly important in order to reduce treatment costs, the complications of urinary infections, and the side effects of medications. In the present study, E. coli was also the most antibiotic-resistant bacteria, especially to fluoroquinolones such as ciprofloxacin and levofloxacin. Therefore, it can be stated that the effect of these antibiotics in the treatment of the E. coli has been decreased.
In this study, a high rate of resistance was observed to ciprofloxacin, levofloxacin, and ceftazidime, respectively; whereas, a low rate of resistance to imipenem, amikacin, and meropenem was also observed. In the study conducted in Arak, a high rate of resistance to cefotaxime, ofloxacin, ciprofloxacin, imipenem, and ceftazidime was reported, respectively (12). Similarly, high resistance rates to ceftazidime and ceftriaxone have been reported from Bandar Abbas, while imipenem and ciprofloxacin were the most active antimicrobial agents against Gram-negative bacilli (13). Additionally, high resistance rates to ceftriaxone were evident from the survey conducted in Khorramabad (14). In the study performed at the Besat Hospital in Tehran, a high resistance rate to ampicillin and the low resistance rate to ampicillin, ceftazidime, amikacin, and cefotaxime was observed (15). The discrepancies observed between these studies and the present study may be the result of the different patient populations, different methodologies, different studied samples, and different common antibiotics used in each hospital.
In general, several studies have been done to investigate the antibiotic resistance in different healthcare settings. In the study conducted by Miller et al. (16), in 2017, less than 10% resistance was reported for the most common Gram-negative bacteria (i.e. Klebsiella spp., Enterobacter spp., and Haemophilus influenza) for ciprofloxacin and/or levofloxacin. The researchers have stated that using fluoroquinolones and aminoglycosides for empirical treatment was the main reason for the observed antibiotic resistance (16). In 2017, Roy et al. (17), conducted a study on 1025 bacteria isolated from the blood samples of children admitted to the hospital of New Delhi to determine the antibiotic resistance pattern. E. coli was the most common isolated Gram-negative bacteria (4.6%) followed by Acinetobacter spp.; Klebsiella spp. showed more than 60% resistance against amikacin and ciprofloxacin. Resistance to third-generation cephalosporines was seen in E. coli, Klebsiella spp., and Enterobactor spp. It can be concluded that antibiotic resistance to these drugs is probably increasing in the E. coli strain and precautions must be taken in their application.
The relatively high frequency of the Pseudomonas spp. strain among the studied urine samples can be attributed to the high frequency of patients with urinary infections. These patients usually received different antibiotic treatments, which may be the bases for the growth of pathogenic bacteria and cause the emergence of bacterial resistance.
Due to the high prevalence of Acinetobacter spp. in the blood and sputum samples establishment of standard precautions and better infection control policies on the education of hospital staff, especially in the ICU, it seems necessary to decrease antibiotic resistance.
This study has some limitations as well. First, this study was conducted in a medical teaching center and the evaluation of the resistance to all of the common antibiotics used for the Gram-negative bacteria was not feasible due to the limited resources. Second, it cannot fully represent the frequency of drug-resistant Gram-negative isolates in Tehran because the study was performed in a limited period of time and a limited number of samples. Thus, future studies with larger samples size would provide better results. Finally, use of broth microdilution susceptibility testing in combination with the disk diffusion method must be considered in future studies.
In conclusion, the relatively high prevalence of resistant pathogens in the current study, merit further attention by health authorities. Establishment of better infection control policies and education of hospital staff, especially in the ICU, are recommended for the prevention and control of drug-resistant pathogens in the health care settings.
References
-
1.
Golsha R, Kazeminejad V, Barzegari A, Besharat S, Ghasemi Kebria F. Antibiotic resistance pattern of Gram-negative bacteria in Gorgan. Med Lab J. 2014;7(5):71-4.
-
2.
Gunseren F, Mamikoglu L, Ozturk S, Yucesoy M, Biberoglu K, Yulug N, et al. A surveillance study of antimicrobial resistance of Gram-negative bacteria isolated from intensive care units in eight hospitals in Turkey. J Antimicrob Chemother. 1999;43(3):373-8. [PubMed ID: 10223593]. https://doi.org/10.1093/jac/43.3.373.
-
3.
Tacconelli E, Carrara E, Savoldi A, Harbarth S, Mendelson M, Monnet DL, et al. Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect Dis. 2018;18(3):318-27. [PubMed ID: 29276051]. https://doi.org/10.1016/S1473-3099(17)30753-3.
-
4.
Archibald L, Phillips L, Monnet D, McGowan JJ, Tenover F, Gaynes R. Antimicrobial resistance in isolates from inpatients and outpatients in the United States: Increasing importance of the intensive care unit. Clin Infect Dis. 1997;24(2):211-5. [PubMed ID: 9114149]. https://doi.org/10.1093/clinids/24.2.211.
-
5.
Cleven BE, Palka-Santini M, Gielen J, Meembor S, Kronke M, Krut O. Identification and characterization of bacterial pathogens causing bloodstream infections by DNA microarray. J Clin Microbiol. 2006;44(7):2389-97. [PubMed ID: 16825354]. [PubMed Central ID: PMC1489523]. https://doi.org/10.1128/JCM.02291-05.
-
6.
Hsueh PR, Chen ML, Sun CC, Chen WH, Pan HJ, Yang LS, et al. Antimicrobial drug resistance in pathogens causing nosocomial infections at a university hospital in Taiwan, 1981-1999. Emerg Infect Dis. 2002;8(1):63-8. [PubMed ID: 11749750]. [PubMed Central ID: PMC2730256].
-
7.
Rodriguez-Bano J, Picon E, Gijon P, Hernandez JR, Cisneros JM, Pena C, et al. Risk factors and prognosis of nosocomial bloodstream infections caused by extended-spectrum-beta-lactamase-producing Escherichia coli. J Clin Microbiol. 2010;48(5):1726-31. [PubMed ID: 20181897]. [PubMed Central ID: PMC2863889]. https://doi.org/10.1128/JCM.02353-09.
-
8.
Tenover FC. Mechanisms of antimicrobial resistance in bacteria. Am J Med. 2006;119(6 Suppl 1):S3-10. discussion S62-70. [PubMed ID: 16735149]. https://doi.org/10.1016/j.amjmed.2006.03.011.
-
9.
Gould FK, Brindle R, Chadwick PR, Fraise AP, Hill S, Nathwani D, et al. Guidelines (2008) for the prophylaxis and treatment of methicillin-resistant Staphylococcus aureus (MRSA) infections in the United Kingdom. J Antimicrob Chemother. 2009;63(5):849-61. [PubMed ID: 19282331]. https://doi.org/10.1093/jac/dkp065.
-
10.
Hiramatsu K. Vancomycin-resistant Staphylococcus aureus: A new model of antibiotic resistance. Lancet Infect Dis. 2001;1(3):147-55. [PubMed ID: 11871491]. https://doi.org/10.1016/S1473-3099(01)00091-3.
-
11.
Lowy FD. Antimicrobial resistance: the example of Staphylococcus aureus. J Clin Invest. 2003;111(9):1265-73. [PubMed ID: 12727914]. [PubMed Central ID: PMC154455]. https://doi.org/10.1172/JCI18535.
-
12.
Didgar F, Sarmadian H, Ghasemikhah R. Antimicrobial resistance pattern of Gram–negative bacilli isolated of Vali-Asr Hospital wards in Arak. ISMJ. 2014;17(5):938-47.
-
13.
Karamstegi A, Davodian P, Vaeghi Z. Bacteria isolated from hospital infections and their antibiotic resistance pattern in Bandar Abbas Shahid Mohammadi Hospital. med J Hormozgan Univ. 2000;5(3):14-7.
-
14.
Saleh F, Soleiman Nejad S, Bahrami Chegeni F, Jafari S, Javanmard A, Rouhi S, et al. Determination of bacterial factors causing urinary infections and its antibiotic resistance patterns in patients referred to Khorramabad Hospital, Iran. Pajouhan Sci J. 2018;16(4):1-5. https://doi.org/10.21859/psj.16.4.1.
-
15.
Mohammadimehr M, Feizabadi MM, Bahadori O, Khosravi M. Study of prevalence of Gram-negative bacteria caused nosocomial infections in ICU in Besat Hospital in Tehran and detection of their antibiotic resistance pattern-year 2007. Iran J Med Microbiol. 2009;3(2):47-54.
-
16.
Miller D. Update on the epidemiology and antibiotic resistance of ocular infections. Middle East Afr J Ophthalmol. 2017;24(1):30-42. [PubMed ID: 28546690]. [PubMed Central ID: PMC5433126]. https://doi.org/10.4103/meajo.MEAJO_276_16.
-
17.
Roy MP, Gaind R, Aggarwal KC, Chellani HK, Biswal I. Pattern of pediatric bacterial infection and antibiotic resistance in New Delhi. Indian Pediatr. 2017;54(2):153-4. [PubMed ID: 28285291]. https://doi.org/10.1007/s13312-017-1022-5.