Abstract
Background:
Hepatitis D virus (HDV) is a defective virus dependent on hepatitis B virus (HBV) for its replication. Due to HDV transmission routes, patients undergoing hemodialysis and those with HIV infection are at risk of acquiring HDV.Objectives:
This study was aimed to determine the frequency and genotype of HDV infection among patients with HIV infection and those undergoing hemodialysis.Patients and Methods:
720 cases including 120 patients undergoing hemodialysis, and 600 patients with HIV infection were studied. All cases with positive results for HBsAg were evaluated for the presence of anti-HDV antibodies. Samples with Anti-HDV positive results were subjected to nested PCR for HDV-RNA confirmation, and sequenced for HDV genotype determination.Results:
HBsAg was found in 9 (7.5%) of 120 patients undergoing hemodialysis, and 9 (1.5%) of 600 patients with HIV infection. 3 (33.3%) of patients undergoing hemodialysis with positive results for HBsAg, and 5 (55.5%) of cases with HIV infection and positive results for HBsAg, had positive findings for anti-HDV which were then subjected to nested PCR. The amplification results confirmed that in 3 (37.5%) samples HDV-RNA was detected. Overall 2.5% of patients undergoing hemodialysis, and 0.8% of cases infected with HIV had positive results for anti-HDV and 1.7% and 0.2% of cases undergoing hemodialysis and patients infected with HIV had positive findings for HDV-RNA respectively. All of the HDV isolates were clustered in clade 1.Conclusions:
The survey showed that overall HDV frequency was not high in our high risk cases. Therefore, practitioners and health care managers should become aware of the risk of dual infection with HBV and HDV especially in high risk patients.Keywords
1. Background
Hepatitis D virus (HDV) is a defective satellite virus which its infectivity is dependent on hepatitis B virus (HBV) and affects nearly 20 million people worldwide (1). HDV infection leads to more severe liver disease than HBV monoinfection with accelerated fibrosis progression, earlier hepatic decompensation and an increased risk for the development of hepatocellular carcinoma (2). High prevalence areas for HDV infection include Italy, parts of Eastern Europe, the Amazon basin, Venezuela, Columbia, some Pacific Islands, Pakistan and Western Asia and infection apparently endemic in the Middle East (3-8). Studies from different areas of Iran showed varied prevalence rates of HDV infection from 2.4% to 5.8% in HBV carriers (9, 10). The prevalence of HDV in various high risk groups in Iran was studied in few surveys. (11-14). In Tehran, HDV prevalence was found to be 5.7% in patients with chronic HBV in 2004 (15). Three distinct genotypes and two subtypes are described for HDV. Genotype I is distributed worldwide, but other genotypes appear to be more restricted geographically. Genotype Ι is spread in the United States, the Middle East, and Europe (7, 16). Genotype ΙΙ is predominant in the Far East (17) and Genotype ΙΙΙ is predominantly detected in South America (3). The predominant genotype of HDV reported from Iran is Genotype Ι (18). Recent extensive analysis of HDV sequences has indicated that the various HDV genotypes fall into at least seven clades (19) and in a study (20) proposed an extended classification of the delta virus genus to eight clades. Most studies suggest that most HDV infections are acquired through parenteral and sexual routes (21, 22). Thus patients undergoing hemodialysis and patients infected with HIV are at risk of acquiring HDV. Coinfection with HBV/HDV has been shown to increase the risk of complications and virologic failure in patients with HIV infection (23, 24). Clinical studies regarding the frequency and genotypes of HDV infection in high risk patients were limited in Iran.
2. Objectives
This study aimed to determine the frequency and genotypes of HDV among patients with HIV and those undergoing hemodialysis in Tehran, Iran.
3. Patients and Methods
In this study 720 individuals including 120 patients undergoing hemodialysis and 600 patients with HIV infection as high risk group were enrolled from February to June 2011. The samples were collected from the main dialysis center of Tehran and Iranian Research center for HIV/AIDS in Tehran. This project was approved by Pasteur Institute of Iran ethics committee, and informed consent was obtained from patients prior to their enrollment. All samples were tested for hepatitis B surface antigen (HBsAg), hepatitis C antibody (anti-HCV) and anti-HDV by enzyme-linked immunosorbent assay (ELISA). The commercial enzyme immunoassay kits used were as follows: HBsAg (Hepanosticka Biomerieux, Boxtel, The Netherlands), anti-HCV (Biorad, Segrate, Italy), and anti-HDV (Dia.Pro Diagnostic Bioprobes sri, Milano, Italy). Recombinant immunoblot assay (RIBA Innogenetics, Ghent, Belgium) was employed to confirm anti-HCV reactivity. Human immunodeficiency virus antibody (anti-HIV) was determined by ELISA (MP Biomedicals, Illkirch, France); with positive tests confirmed by Western blot assay (Diaplus, San Francisco, USA). CD4+ count was determined by flowcytometry and defined as cells/mm3. HDV-RNA was extracted from 200 ml of serum using High Pure Viral Nucleic Acid kit (Roche Diagnostics GmbH, Mannheim, Germany) following the manufacturer’s instructions. Then RNA was converted to cDNA, subsequently, cDNA was amplified, using a denaturation step at 95ºC for 3 min, followed by 40 cycles of denaturation at 95ºC for 45 sec, annealing for 45 sec at 61ºC, and extension at 72ºC for 50 sec. RT-PCR products were amplified further using nested PCR (95ºC for 3 min, followed by 35 cycles of 95ºC for 45 sec, 60ºC for 45 sec, and 72ºC for 45 sec) using Taq polymerase (Roche Diagnostics GmbH). The primers used in the RT-PCR/nested PCR were as follows:
E1 (outer; sense), CCA GGT CGG ACC GCG AGG AGG (858–881 bases)
E2 (outer; anti sense), ACA AGG AGA GGC AGG ATC ACC GAC (1312–1289 bases)
E3 (inner; sense), GAT GCC ATG CCG ACC CGA AGA G (883–906 bases)
E4 (inner; anti sense), GAA GGA AGG CCC TCA AGA ACA AGA (1288–1265 bases)
The inner primers amplify a 404 base pair (bp) region in the C-terminal portion of the hepatitis delta antigen (HDAg) coding region and included the RNA editing site and the polyadenylation signal. PCR products were electrophoresed on a 1.5% agarose gel, containing ethidium bromide, and visualized on a gel documentation system. PCR products were purified using the High Pure PCR product purification kit (Roche Diagnostics GmbH, Mannheim, Germany), and sequenced directly at the Sequence Laboratories Gottingen GmbH (SEQLAB), Germany, using the inner primers. Sequences were compared with a set of published sequences corresponding to all the HDV clades shown in Figure 1.
Phylogenetic Tree Constructed Using a Kimura Two-Parameter Matrix and the Neighbor-Joining Method of HDV Strains
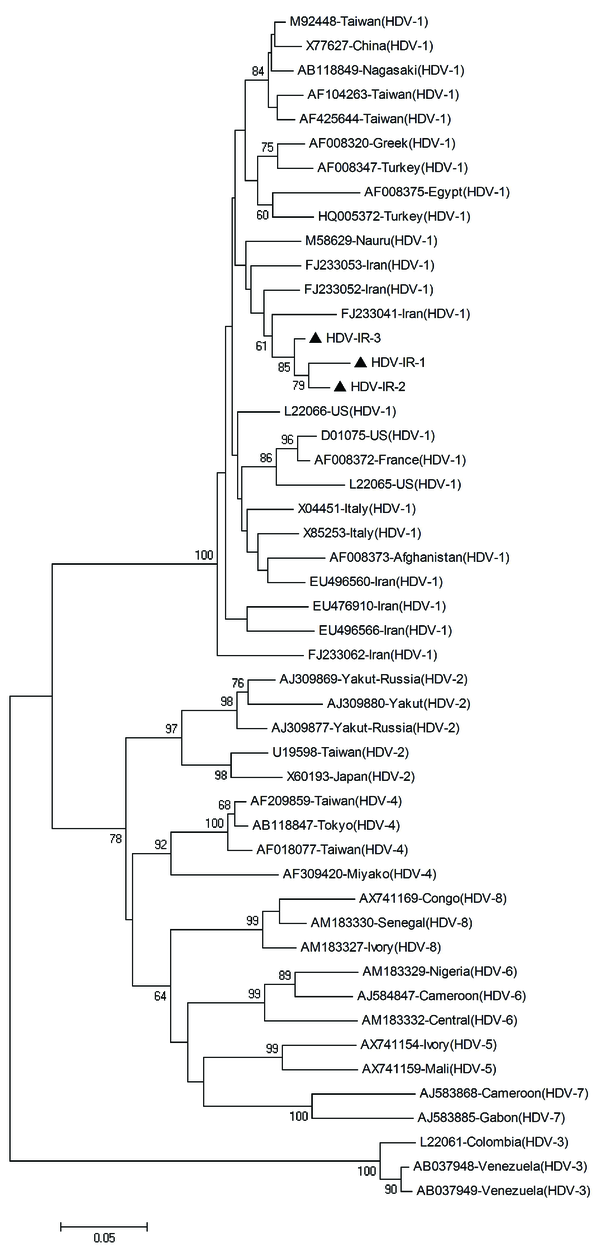
Phylogenetic and molecular evolutionary analyses were conducted using MEGA version 4 (25) for phylogeny and BioEdit (The BioEdit Sequence Alignment Editor software, Department of the Microbiology, North California State University) for sequencing analysis. Nucleotide sequences were aligned with CLUSTALW program and confirmed by visual inspection. The Kimura two-parameter algorithm was used for genetic distance calculation (26). A phylogenetic tree was generated by the neighbor-joining method, and bootstrap resampling and reconstruction was performed 1000 × to confirm the reliability of the phylogenetic tree (27).
The HDV sequences used in this study were deposited in GenBank under accession numbers JF694492 through JF694494.
3.1. Statistical Analysis
The Chi-square was used with the SPSS 16 Package program for statistical analysis (Chicago, IL, USA). Data were presented as mean ± SD or, when indicated, as an absolute number and percentage.
4. Results
In this study 720 individuals including 120 patients undergoing hemodialysis and 600 patients with HIV infection were enrolled. The mean age of patients with HIV was 36.9 ± 9.2 (range: 9-67) years. 69.6% of patients were male and 30.4% were female. The mean CD4 + count of patients was 275.6 ± 181 (16-1000) cells/mm3. The presumed routes of HIV transmission were intravenous drug use (53.6%), heterosexual contact (28.7%), infected blood and blood products transfusion (2.8%), vertical transmission (1.1%), tattooing (0.6%), injection drug use and tattooing (0.6%), heterosexual contact and intravenous drug use (4.4%), heterosexual contact and infected blood (0.6%), and in 7.6% the route of HIV acquisition was not identified. The mean age of patients undergoing hemodialysis was 55 ± 16 (range: 15-89) years. 60% of patients were male and 40% were female. The duration of hemodialysis was 5.2 ± 5.1 years. All of them had past history of blood transfusion, but none of them had organ transplantation. HBsAg was found in 9 (7.5%) of 120 patients undergoing hemodialysis and 9 (1.5%) of 600 patients with HIV infection. 3 (33.3%) of patients undergoing hemodialysis with positive results for HBsAg and 5 (55.5%) of cases with HIV infection and positive results for HBsAg, had positive findings for anti-HDV which were then subjected to nested PCR. The amplification results confirmed that in 3 (37.5%) samples HDV-RNA was detected. Two of them were under hemodialysis and one was HIV case. All HDV infected cases were male. All of the subjects coinfected with HIV/HBV/HDV (serology positive) were intravenous drug users, and were coinfected with HCV. Overall 2.5% of patients undergoing hemodialysis, and 0.8% of patients infected with HIV had positive results for anti-HDV, and 1.7% and 0.2% of cases undergoing hemodialysis and patients infected with HIV had positive findings for HDV-RNA respectively. Frequency of HBV and HDV infection in cases undergoing hemodialysis, and patients infected with HIV was summarized in Table 1.
Prevalence of HBV and HDV Infection inPatients Undergoing Hemodialysis and Those with HIV Infection
| Cases Characteristic | Patients undergoing Hemodialysis (n=120) | Cases Characteristic (n=600) |
|---|---|---|
| Sex, Male/Female | 72/48 | 418/182 |
| Age, mean ± SD, y | 55±16 | 36.9±9.2 |
| HBsAg(+)a, No. (%) | 9 (7.5%) | 9 (1.5%) |
| Anti-HDV(+), No. (%) | 3 (2.5%) | 5 (0.83%) |
| HDV-RNA(+), No. (%) | 2 (1.66%) | 1 (0.16%) |
Phylogenetic analysis revealed that all HDV isolates were classified as clade 1 (genotype I) (Figure. 1).
5. Discussion
The prevalence of HDV is variable, predominating in some areas of the world (28). Most studies suggest that most HDV infections are acquired through parenteral and sexual routes (21, 22), thus cases undergoing hemodialysis and patients infected with HIV are at risk of acquiring HDV. The prevalence of HDV infection in patients infected with HIV and HBV ranges from 1.2% in the southeast region of Brazil to 14.5% in south and/or east Europe, and 22.2% in Taiwan (29-31). Coinfection is especially common among patients who are injection drug users (23, 24). Fainboim et al. (23) showed a low prevalence of anti-HDV in patients infected with HIV (1.9%). Anti-HDV was significantly higher in injection drug users (3.4%) than heterosexuals (0%). There was no significant difference in the percentage of anti-HDV positivity between injection drug users and homosexuals (1.0%). In another investigation, 3.9% of cases had triple infection with HIV/HBV/HDV (32). In a recent survey in Taiwan, 22.2% of patients infected with HIV with chronic HBV coinfection had positive findings for anti-HDV (30). A recently published study in Iran has shown a high HDV prevalence of 31.57% in individuals with HIV/HBV coinfection in Kermanshah, Western Iran (33). The current study showed that 1.5% of cases with HIV infection had HBV infection. It can be due to increased knowledge about HBV transmission routes, national vaccination program, and HBV vaccination of high risk patients. 0.8% of patients infected with HIV are coinfected with both HBV and HDV. All of the subjects coinfected with HIV/HBV/HDV were intravenous drug users. On the other hand, higher frequency of HDV infection (55.5%) was found among patients with HBV/HIV coinfection than that in the general population of Iranian HBsAg carriers (2.4-5.8%). The prevalence of HDV infection in patients with chronic hemodialysis is not clear (34). In studies by Ramia (35) and Voiculescu et al. (36), anti-HDV was not found in patients undergoing hemodialysis. Abraham et al. (37) reported that 2.9% of renal transplant recipients were infected with HDV. In an investigation in Iran, 44.5% of patients undergoing hemodialysis with HBV infection were infected with HDV (14). In another study in this country, 25.2% of patients undergoing hemodialysis and positive results for HBsAg were found to be anti-HDV positive (13). The findings showed that overall 33.3% of patients undergoing hemodialysis in Tehran had positive findings for anti-HDV which is similar to other reports from Iran. The results reveal relatively high frequency of HDV infection in cases coinfected with HIV/HBV (55.5%) and patients undergoing hemodialysis with positive results for HBsAg (33.3%), although overall HDV frequency might not be high in high risk groups. Recently, eight major clades of HDV (HDV-1 to HDV-8) were described. HDV-1 and HDV-3 are distributed worldwide, but other genotypes appear to be more restricted geographically. HDV-2 is found in Japan, Taiwan, and Yakutia, Russia; HDV-4 in Taiwan and Japan and HDV-5, -6, -7, -8 in Africa (20). The study revealed HDV clade I (genotype 1) in all of the patients. This result is in agreement with predominant genotype of HDV reported in other studies from Iran (18, 38), and also the Middle East region such as Lebanon Egypt, Turkey and Pakistan (17, 35, 39, 40). Previous findings indicated that there may be an association between the geographical distribution of HDV and HBV genotypes (38). As all reports from Iran showing that genotype D is the only detectable genotype in all clinical forms of HBV infection in this country (41, 42). It is expected that HDV genotype I was detected as predominant genotype in this study. In conclusion, this survey showed, although overall HDV frequency might not be high in the cases, an increase in HDV occurrence in the cohort of high risk patients were seen. Therefore, practitioners and health care managers should become aware of the risk of dual infection with HBV and HDV especially in high risk patients.
Acknowledgements
References
-
1.
Deny P. Hepatitis delta virus genetic variability: from genotypes I, II, III to eight major clades? Curr Top Microbiol Immunol. 2006;307:151-71. [PubMed ID: 16903225].
-
2.
Grabowski J, Wedemeyer H. Hepatitis delta: immunopathogenesis and clinical challenges. Dig Dis. 2010;28(1):133-8. [PubMed ID: 20460901]. https://doi.org/10.1159/000282076.
-
3.
Casey JL, Brown TL, Colan EJ, Wignall FS, Gerin JL. A genotype of hepatitis D virus that occurs in northern South America. Proc Natl Acad Sci U S A. 1993;90(19):9016-20. [PubMed ID: 8415646].
-
4.
Anonymous. Centers for Disease Control. USA: MMWR; 1990.Hepatitis Surveillance: Report. 23 p.
-
5.
Gaeta GB, Stroffolini T, Chiaramonte M, Ascione T, Stornaiuolo G, Lobello S, et al. Chronic hepatitis D: a vanishing Disease? An Italian multicenter study. Hepatology. 2000;32(4 Pt 1):824-7. [PubMed ID: 11003629]. https://doi.org/10.1053/jhep.2000.17711.
-
6.
Mumtaz K, Hamid SS, Adil S, Afaq A, Islam M, Abid S, et al. Epidemiology and clinical pattern of hepatitis delta virus infection in Pakistan. J Gastroenterol Hepatol. 2005;20(10):1503-7. [PubMed ID: 16174065]. https://doi.org/10.1111/j.1440-1746.2005.03857.x.
-
7.
Rizzetto M, Hadziyannis S, Hannson B, Toukan A, Gust L, Amoroso P, et al. Hepatitis delta virus infection in the world: epidemiological patterns and clinical expression. Gastroenterol Inte. 1992;5(1):18-32.
-
8.
Rizzetto M, Ponzetto A, Forzani I. Epidemiology of hepatitis delta virus: overview. Prog Clin Biol Res. 1991;364:1-20. [PubMed ID: 2020686].
-
9.
Bhattacharyya S, Dalal BS, Lahiri A. Hepatitis D infectivity profile among hepatitis B infected hospitalised patients in Calcutta. Indian J Public Health. 1998;42(4):108-12. [PubMed ID: 10389522].
-
10.
Jaiswal SP, Chitnis DS, Artwani KK, Naik G, Jain AK. Prevalence of anti-delta antibodies in central India. Trop Gastroenterol. 1999;20(1):29-32. [PubMed ID: 10464445].
-
11.
Abbas Z, Jafri W, Raza S. Hepatitis D: Scenario in the Asia-Pacific region. World J Gastroenterol. 2010;16(5):554-62. [PubMed ID: 20128022].
-
12.
Amini N, Alavian SM, Kabir A, Saiedi Hosseini SY, Aalaei Andabili SH. Clinical Features and Seroepidemiology of Anti-HDV Antibody in patients With Chronic Hepatitis B Virus Infection in Iran: A Meta-Analysis. Hepat Mon. 2011;11(12):960-7. [PubMed ID: 22368679]. https://doi.org/10.5812/kowsar.1735143X.805.
-
13.
KARIMI A, AMINI SAFIEH AMIRKHANI AA. Investigation and Comparison of hepatitis D prevalence in dialysis patients and the donors of HBsAg carrier. Teb Va Tazkieh. 2000;36(30):3.
-
14.
Rezvan H, Forouzandeh B, Taroyan S, Fadaiee S, Azordegan F. A study on delta virus infection and its clinical impact in Iran. Infection. 1990;18(1):26-8. [PubMed ID: 2155876].
-
15.
Alavian SM, Assari S, Manzoori-Joybari H, MoghaniLankarani M, Doroudi T, Haji-Beigi B, et al. Frequency and risk factors of hepatitis D virus in hepatitis B patients. Govaresh. 2012;10(1):21-6.
-
16.
Shakil AO, Hadziyannis S, Hoofnagle JH, Di Bisceglie AM, Gerin JL, Casey JL. Geographic distribution and genetic variability of hepatitis delta virus genotype I. Virology. 1997;234(1):160-7. [PubMed ID: 9234957]. https://doi.org/10.1006/viro.1997.8644.
-
17.
Moatter T, Abbas Z, Shabir S, Jafri W. Clinical presentation and genotype of hepatitis delta in Karachi. World J Gastroenterol. 2007;13(18):2604-7. [PubMed ID: 17552010].
-
18.
Mohebbi SR, Zali N, Derakhshan F, Tahami A, Mashayekhi R, Amini-Bavil-Olyaee S, et al. Molecular epidemiology of hepatitis delta virus (HDV) in Iran: a preliminary report. J Med Virol. 2008;80(12):2092-9. [PubMed ID: 19040284]. https://doi.org/10.1002/jmv.21326.
-
19.
Radjef N, Gordien E, Ivaniushina V, Gault E, Anais P, Drugan T, et al. Molecular phylogenetic analyses indicate a wide and ancient radiation of African hepatitis delta virus, suggesting a deltavirus genus of at least seven major clades. J Virol. 2004;78(5):2537-44. [PubMed ID: 14963156].
-
20.
Le Gal F, Gault E, Ripault MP, Serpaggi J, Trinchet JC, Gordien E, et al. Eighth major clade for hepatitis delta virus. Emerg Infect Dis. 2006;12(9):1447-50. [PubMed ID: 17073101]. https://doi.org/10.3201/eid1209.060112.
-
21.
Liaw YF, Chiu KW, Chu CM, Sheen IS, Huang MJ. Heterosexual transmission of hepatitis delta virus in the general population of an area endemic for hepatitis B virus infection: a prospective study. J Infect Dis. 1990;162(5):1170-2. [PubMed ID: 2121838].
-
22.
Stroffolini T, Ferrigno L, Cialdea L, Catapano R, Palumbo F, Novaco F, et al. Incidence and risk factors of acute Delta hepatitis in Italy: results from a national surveillance system. SEIEVA Collaborating Group. J Hepatol. 1994;21(6):1123-6. [PubMed ID: 7699238].
-
23.
Fainboim H, Gonzalez J, Fassio E, Martinez A, Otegui L, Eposto M, et al. Prevalence of hepatitis viruses in an anti-human immunodeficiency virus-positive population from Argentina. A multicentre study. J Viral Hepat. 1999;6(1):53-7. [PubMed ID: 10847130].
-
24.
Shukla NB, Poles MA. Hepatitis B virus infection: co-infection with hepatitis C virus, hepatitis D virus, and human immunodeficiency virus. Clin Liver Dis. 2004;8(2):445-60. [PubMed ID: 15481349].
-
25.
Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24(8):1596-9. [PubMed ID: 17488738]. https://doi.org/10.1093/molbev/msm092.
-
26.
Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16(2):111-20. [PubMed ID: 7463489].
-
27.
Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4(4):406-25. [PubMed ID: 3447015].
-
28.
Solomon RE, Kaslow RA, Phair JP, Lyter D, Visscher B, Lyman D, et al. Human immunodeficiency virus and hepatitis delta virus in homosexual men. A study of four cohorts. Ann Intern Med. 1988;108(1):51-4. [PubMed ID: 3337516].
-
29.
Mendes-Correa MC, Gomes-Gouvea MS, Alvarado-Mora MV, Da Silva MH, Lazari C, Cavalcanti NC, et al. Hepatitis delta in HIV/HBV co-infected patients in Brazil: is it important? Int J Infect Dis. 2011;15(12):e828-32. [PubMed ID: 21999909]. https://doi.org/10.1016/j.ijid.2011.08.003.
-
30.
Sheng WH, Hung CC, Kao JH, Chang SY, Chen MY, Hsieh SM, et al. Impact of hepatitis D virus infection on the long-term outcomes of patients with hepatitis B virus and HIV coinfection in the era of highly active antiretroviral therapy: a matched cohort study. Clin Infect Dis. 2007;44(7):988-95. [PubMed ID: 17342655]. https://doi.org/10.1086/511867.
-
31.
Soriano V, Grint D, d'Arminio Monforte A, Horban A, Leen C, Poveda E, et al. Hepatitis delta in HIV-infected individuals in Europe. AIDS. 2011;25(16):1987-92. [PubMed ID: 21857493]. https://doi.org/10.1097/QAD.0b013e32834babb3.
-
32.
Boyd A, Lacombe K, Miailhes P, Gozlan J, Bonnard P, Molina JM, et al. Longitudinal evaluation of viral interactions in treated HIV-hepatitis B co-infected patients with additional hepatitis C and D virus. J Viral Hepat. 2010;17(1):65-76. [PubMed ID: 19682317]. https://doi.org/10.1111/j.1365-2893.2009.01153.x.
-
33.
Vaziri S, Mansouri F, Sayad B, Afsharian M, Janbakhsh A, Karami M. Hepatitis D virus infection among HIV-HBV co-infected patients in Kermanshah, West of Iran. Hepat Mon. 2008;8:252-257.
-
34.
Zuckerman M. Surveillance and control of blood-borne virus infections in haemodialysis units. J Hosp Infect. 2002;50(1):1-5. [PubMed ID: 11825044]. https://doi.org/10.1053/jhin.2001.1068.
-
35.
Ramia S, El-Zaatari M, Sharara AI, Ramlawi F, Farhat B. Current prevalence of hepatitis delta virus (HDV) infection and the range of HDV genotypes in Lebanon. Epidemiol Infect. 2007;135(6):959-62. [PubMed ID: 17176498]. https://doi.org/10.1017/S0950268806007643.
-
36.
Voiculescu M, Iliescu L, Ionescu C, Micu L, Ismail G, Zilisteanu D, et al. A cross-sectional epidemiological study of HBV, HCV, HDV and HEV prevalence in the SubCarpathian and South-Eastern regions of Romania. J Gastrointestin Liver Dis. 2010;19(1):43-8. [PubMed ID: 20361074].
-
37.
Abraham P, John GT, Raghuraman S, Radhakrishnan S, Thomas PP, Jacob CK, et al. GB virus C/hepatitis G virus and TT virus infections among high risk renal transplant recipients in India. J Clin Virol. 2003;28(1):59-69. [PubMed ID: 12927752].
-
38.
Mirshafiee H, Mahmoodian-Shooshtari M, Sharifi Z, Hosseini SM. Genotype analysis of hepatitis delta virus from hepatitis B surface antigen-positive patients using PCR-RFLP in Tehran, Iran. Arch Iran Med. 2009;12(3):238-43. [PubMed ID: 19400600].
-
39.
Bozdayi AM, Aslan N, Bozdayi G, Turkyilmaz AR, Sengezer T, Wend U, et al. Molecular epidemiology of hepatitis B, C and D viruses in Turkish patients. Arch Virol. 2004;149(11):2115-29. [PubMed ID: 15503201]. https://doi.org/10.1007/s00705-004-0363-2.
-
40.
Saudy N, Sugauchi F, Tanaka Y, Suzuki S, Aal AA, Zaid MA, et al. Genotypes and phylogenetic characterization of hepatitis B and delta viruses in Egypt. J Med Virol. 2003;70(4):529-36. [PubMed ID: 12794714]. https://doi.org/10.1002/jmv.10427.
-
41.
Aghakhani A, Hamkar R, Zamani N, Eslamifar A, Banifazl M, Saadat A, et al. Hepatitis B virus genotype in Iranian patients with hepatocellular carcinoma. Int J Infect Dis. 2009;13(6):685-9. [PubMed ID: 19138545]. https://doi.org/10.1016/j.ijid.2008.10.009.
-
42.
Mojiri A, Behzad-Behbahani A, Saberifirozi M, Ardabili M, Beheshti M, Rahsaz M, et al. Hepatitis B virus genotypes in southwest Iran: molecular, serological and clinical outcomes. World J Gastroenterol. 2008;14(10):1510-3. [PubMed ID: 18330939].