Abstract
Background:
Mutations in basal core promoter (BCP) and precore regions of hepatitis B virus (HBV) are associated with course and treatment outcomes of chronic HBV infection. While BCP and precore mutation analysis have been carried out in adult patients between different genotypes, this analysis has rarely been performed for chronically infected children.Objectives:
The aim of this study was to assess the mutation profiles of BCP and precore regions in different HBV genotypes in chronically infected children.Patients and Methods:
A cohort of 245 children and 92 adults with chronic HBV infection was included in this study. BCP and precore regions were analyzed by PCR amplification and sequenced.Results:
Ten nucleotide positions, including nt1679, nt1721, nt1753, nt1757, nt1758, nt1762, nt1764, nt1775, nt1856 and nt1858 in BCP/precore regions of HBV genome, showed obviously higher frequencies of mutation in genotype C subjects than in genotype B subjects among children, while there were only three positions, including nt1679, nt1758 and nt1775 showing higher mutation frequencies in genotype C subjects than in genotype B subjects in adults. Several combined mutations were obviously highly distributed in children with chronic HBV genotype C infection, such as G1721A/A1775G/T1858C triple mutation; a novel combined mutation type, exclusively detected in children with chronic HBV genotype C infection. In addition, G1721A/A1775G/T1858C combined mutation was associated with higher viral load and lower age distribution.Conclusions:
The mutation ratio difference between genotypes B and C in children was higher than that of adults and several combined mutations were exclusively detected in children with chronic HBV genotype C infection associated with higher viral load.Keywords
1. Background
Hepatitis B virus (HBV) infection is a global public health problem. Approximately 240 million people globally are chronically infected with HBV (1). The spectrum of the symptomatology of HBV infection varies in both acute and chronic phases. During the acute phase, manifestations range from subclinical or anicteric hepatitis to icteric hepatitis and in some cases fulminant hepatitis. During the chronic phase, symptoms range from an asymptomatic carriers state to chronic hepatitis, cirrhosis, and hepatocellular carcinoma (HCC). Pathogenesis and the clinical outcome of HBV infection is determined by a complex interaction between viral factors and host immune responses (2).
The pathogenesis and treatment outcomes of chronic HBV infections, including hepatitis B e antigen (HBeAg) clearance, liver damage, response to interferon (IFN) treatment and the incidence of HCC, are associated with certain mutations and the genotype of the virus. Based on a sequence divergence of > 8% of HBV genome, 10 genotypes (A-J) have been identified so far (3, 4). Genotypes B and C are endemic genotypes in China (5). Patients with genotype C tend to have a longer HBeAg carrier status and a higher prevalence compared with patients with genotype B (6, 7). Genotype C HBV is more prevalent in patients with cirrhosis and has a higher histologic activity score (8, 9). Genotype C is associated with a lower response rate to conventional interferon therapy compared with genotype B (10). Infection with genotype C is often associated with the risks of liver cirrhosis and HCC at older ages compared with infection with genotype B (9, 11, 12).
Basal core promoter (BCP), regulated by enhancer II and to some extent by enhancer I, controls the transcription of precore mRNA and pregenomic RNA (13). HBeAg is encoded by the precore region of HBV genome; it is an indicator for active viral replication and plays an immune regulatory role during infection (14-16). Mutations in precore region of HBV are associated with high viral load (17). Most of the BCP/precore mutation analyses have been performed in adult patients. Compared with adult patients, children have always been in the immune tolerance phase, presenting a high ratio of HBeAg positivity and high levels of serum HBV DNA compared to normal or minimally elevated serum alanine aminotransferases (ALT). Previous studies have predominantly examined specific viral mutations alone, such as nt1896 and nt1899, and rarely investigated the presence of other mutations. In addition, the "wild-type" patterns of the reference HBV genotype have been chosen randomly, introducing some controversial results.
2. Objectives
In this study, we initially identify the wild-type nucleotides in the BCP/precore regions of HBV genotypes B and C from the HBV sequences obtained from NCBI. HBV sequences from 245 children (including 191 genotype B and 54 genotype C sequences) and 92 adults (including 66 genotype B and 26 genotype C sequences) were also evaluated for mutation discrepancies of the BCP/precore regions.
3. Patients and Methods
3.1. Study Subjects
A total of 337 serum samples were obtained from a cohort of patients with chronic HBV infection, including 245 children and 92 adults. All of these patients had visited the Children’s Hospital of Chongqing Medical University, the first affiliated hospital of Chongqing Medical University, the second affiliated hospital of Chongqing Medical University, Guangzhou Women and Children’s Medical Center and Guangdong Women and Children’s Hospital between June 2011 and September 2013. All the samples were stored at −70°C until further testing. All the patients were positive for hepatitis B surface antigen (HBsAg), but negative for anti-HBs, anti-hepatitis C virus (HCV), anti-hepatitis D virus (HDV) and anti-human immunodeficiency virus (HIV). They did not receive any antiviral or immunosuppressive therapy before samples collection. Informed written consents were obtained from all the patients or their parents. This study was approved by the Ethical Review Committee for Research of Chongqing Medical University.
3.2. Hepatitis B Virus DNA Amplification and Sequencing
The HBV viral DNA was extracted from 200 µL of patient serum samples using the QIAamp DNA blood mini kit (Qiagen, Hilden, Germany) and eluted with 100 µL elusion buffer according to the manufacturers’ instructions. The precore and core promoter regions (nt1653-1959) of the HBV genome were amplified by nested PCR with PrimeSTAR HS DNA polymerase (Premix, Takara), as previously studied (18). The amplified PCR products were sequenced by ABI 3100 genetics analyzer. The complete S gene (nt65 to 870) of 337 samples was amplified by nested PCR, as described previously (19), and then analyzed using the NCBI genotyping tool (http://www.ncbi.nlm.nih.gov/projects/genotyping/formpage.cgi) to identify the genotype of a viral sequence.
3.3. Definition of Wild-Type Nucleotides of Genotypes B and C Sequences and Mutation Analysis
A total of 784 HBV genotype B sequences and 1289 HBV genotype C sequences were downloaded from the NCBI GenBank. After sequences alignment, a nucleotide with the highest frequency at each position in the BCP/precore regions of HBV genotype B or genotype C sequences was termed as a wild-type nucleotide. Nucleotide substitutions with other three nucleotides at each position were termed as mutations.
3.4. Statistical Analysis
Chi-squared test and Fisher’s exact test were used to determine the differences between the groups in categorical variables such as frequency of mutation. Wilcoxon-Mann-Whitney test was used for continuous variables such as ALT level, DNA load and the number of nucleotide mutations. P < 0.05 was considered statistically significant. All the tests were analyzed by SPSS version 17.0 software (IBM, USA).
4. Results
4.1. Clinical and Viral Characteristics of Subjects
A total of 245 chronic HBV infected children and 92 infected adults were included in the present study (Table 1). For For the children group, 191 HBV sequences belonged to genotype B and the remaining 54 sequences belonged to genotype C. The distribution of sequences in adults was 66:26 for genotype B to C, respectively. Genotype B sequences were the major components for both children and adults groups. There were no significant differences in ALT levels, DNA load, HBeAg-positive ratio, sexual distinction and age between patients with genotypes B and C.
| Children | Adults | |||||
|---|---|---|---|---|---|---|
| Genotype B (n = 191) | Genotype C (n = 54) | P Value | Genotype B (n = 66) | Genotype C (n = 26) | P Value | |
| Gender | 0.171 | 0.938 | ||||
| Male | 121 | 28 | 48 | 18 | ||
| Female | 70 | 26 | 18 | 8 | ||
| Age, y | 6.72 ± 4.08 | 5.69 ± 3.83 | 0.098 | 38.35 ± 9.57 | 40.31 ± 8.77 | 0.323 |
| HBeAg(+)/HBeAg(-) | 116/75 | 39/15 | 0.166 | 31/35 | 13/13 | 0.976 |
| ALT, IU/L | 57.7 ± 89.4 | 106.4 ± 119.9 | 0.089 | 139.2 ± 116.1 | 136.3 ± 106.3 | 0.775 |
| DNA loading, log copies/mL | 6.57 ± 2.28 | 6.91 ± 2.44 | 0.056 | 6.68 ± 1.64 | 6.01 ± 1.67 | 0.058 |
4.2. Nucleotides in Basal Core Promoter/Precore Regions With Varied Mutation Ratios in Different Hepatitis B Virus Genotypes
Table 2 displays various nucleotide mutations and frequencies within genotypes B and C sequences among adults and children groups. In children, the frequencies of mutations at nucleotides1679, 1721, 1753, 1757, 1758, 1762, 1764, 1775, 1856 and 1858 were significantly higher in genotype C compared with genotype B strains. The opposite was true for mutation at nt1752. For mutation at nt1721, for example, the wild-type nucleotide at this position was “G” in both genotypes C and B sequences and the mutation type at this position was “A” for both genotypes sequences. However, the frequency of G1721A mutation in genotype C strains was higher than genotype B (31.5% vs 0.5%, P < 0.001). Similar mutation patterns were also observed at nt1775 (29.6% for genotype C and 1.6% for genotype B, P < 0.001), and at nt1856 and nt1858 those were only observed in genotype C sequences (16.7% and 31.5%, respectively). On the contrary, the mutation at nt1752 was more popular in genotype B than in genotype C strains (46.1% vs 5.6%, P < 0.001).
Nucleotides with different mutation frequencies between genotypes B and C were also observed at nt1679, 1752, 1758, and 1775 in HBV genomes isolated from adults. Mutations of nt1679, 1758 and 1775 predominantly occurred in patients with genotype C, while the nt1752 mutation presented the opposite characteristics. Partial hotspot mutation positions (including nt1721, 1753, 1757, 1762, 1764, 1856 and 1858) found in children were not observed in adult patients. G1896 and G1899 mutations were associated with “e” antigen status, as reported in a previous study (20), but mutation frequencies of these two sites were not significantly different between genotypes B and C. As a result, these two sites were not included for further analysis.
The proportion of HBeAg-positive samples in children with genotype B infection was not significantly different from those with genotype C infection (60.7% vs 72.2%, P > 0.05). The observed nucleotides with different mutation frequencies between genotypes B and C sequences may not be attributed to the “e” antigen status of the patients. Similar conclusion can also be applied to adults (46.97% vs 50%, P > 0.05).
Frequencies of Mutations in Basal Core Promoter/Precore Regions Correlated With Different Hepatitis BVirus Genotypes
| Mutation Type | Children | Adults | ||||
|---|---|---|---|---|---|---|
| Genotype B (n = 191) | Genotype C (n = 54) | P Value | Genotype B (n = 66) | Genotype C (n = 26) | P Value | |
| A1679G | 1.6 | 14.8 | < 0.001 | 0.0 | 11.5 | 0.026 |
| G1721A | 0.5 | 31.5 | < 0.001 | 3.0 | 3.8 | 0.636 |
| A1752G | 46.1 | 5.6 | < 0.001 | 53.0 | 3.8 | < 0.001 |
| T1753G/C | 0.5 | 5.6 | 0.035 | 1.5 | 11.5 | 0.120 |
| G1757A | 1.6 | 16.7 | < 0.001 | 3.0 | 3.8 | 1.0 |
| T1758C | 0.5 | 18.5 | < 0.001 | 1.5 | 19.2 | 0.009 |
| A1762T | 16.2 | 27.8 | 0.009 | 24.2 | 38.5 | 0.173 |
| G1764A | 16.2 | 27.8 | 0.009 | 24.2 | 38.5 | 0.173 |
| A1775G | 1.6 | 29.6 | < 0.001 | 1.5 | 23.1 | 0.002 |
| C1856T | 0.0 | 16.7 | < 0.001 | 0.0 | 0.0 | 1.0 |
| T1858C | 0.0 | 31.5 | < 0.001 | 0.0 | 3.8 | 0.627 |
| G1896A | 18.3 | 9.3 | 0.112 | 57.6 | 46.2 | 0.322 |
| G1899A | 2.6 | 5.6 | 0.379 | 10.6 | 15.4 | 0.498 |
4.3. Number of Nucleotide Mutations in Basal Core Promoter/Precore Regions of Hepatitis BVirus
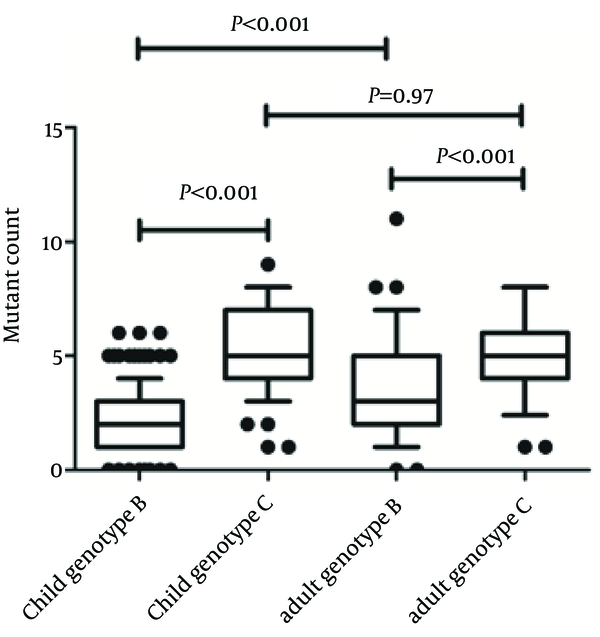
The number of nucleotide mutations within the BCP and precore regions were different between HBV genotypes C and B strains. The average number of nucleotide mutations in this region in genotype C strains was significantly higher than in genotype B strains, in both children and adults (5.17 ± 1.92 vs 2.58 ± 2.24 in children, P < 0.001; 5.08 ± 1.83 vs 3.50 ± 2.21 in adults, P < 0.001, Figure 1). In addition, we compared the average number of mutations in BCP/precore region of HBV genotypes in different age groups. We found that the average number of nucleotide mutations in children with genotype B was obviously lower than that of adults with genotype B (2.58 ± 2.24 vs 3.50 ± 2.21, P < 0.001), while there was no difference between genotype C strains (5.17 ± 1.92 vs 5.08 ± 1.83, P = 0.97, Figure 1). This result suggests that older patients infected with HBV genotype B strain possess a higher virus sequence complexity.
The Number of Average Nucleotides Mutation in Basal Core Promoter/Precore Regions in Genotype B/C Children and Adults
4.4. Combined Mutations in Basal Core Promoter/Precore Regions Associated With Different Hepatitis B Virus Genotypes
Nucleotides with different mutation frequencies associated with different genotypes were predominantly detected in combination with each other, particularly in genotype C sequences from children with HBV infection (Table 3). For example, the frequency of T1858C mutation in genotype C sequences from children was 31.5% (17/54). This mutation was detected in combination with other mutations, as exemplified by the G1721A/A1775G/T1858C triple-mutation (16/17, 94.1%). These combined mutations were not detected in genotype B sequences from children adults. Similarly, most of the C1856T mutations from genotype C sequences of the HBV-infected children were also detected in combined mutation forms (G1721A/A1775G/C1856T/T1858C, 8/9, 88.9%). Combined mutation characteristics were also found in other hotspot mutation positions from genotype C sequences of children, such as G1721A (G1721A/A1775G/T1858C, 16/18) and A1775G mutations (G1721A/A1775G/T1858C, 16/19). Therefore, our results showed that combined mutations were mainly found in genotype C sequences from children with chronic HBV infection.
4.5. The Association Between Combined Mutations and Clinical Characters in Children
The association between 19 combined mutations and clinical characteristics, including viral DNA titer, ALT level, age and HBeAg status, were analyzed in children with HBV infection. Most combined mutations had no correlation with clinical phenotypes, with exception to the G1721A/A1775G/T1858C triple mutation. This triple mutation was significantly correlated with the genotype C virus sequences, lower age and DNA load. The age of children containing G1721A/A1775G/T1858C triple-mutation strains was significantly lower than that of triple-mutation-free patients (4.56 ± 2.45 vs 6.56 ± 4.12, P = 0.012, Table 4). Similarly, the dominant genotype of the triple-mutation-containing patients was genotype C (16/16), while the main genotype in the without the triple-mutation group was genotype B (191/229, P < 0.001). The DNA load of the triple-mutation-containing patients was significantly higher than that of the triple-mutation-free group (7.56 ± 2.07 vs 6.64 ± 2.29, P = 0.045, Table 4). Despite the lack of statistically significant difference, the ALT value of the triple-mutation-containing subjects tended to be higher than the triple-mutation-free subjects (70.7 ± 62.2 vs 51.5 ± 36.9,P = 0.298, Table 4). There were no significant differences in frequency of this triple-mutation between HBeAg positive and HBeAg negative children (8.4% vs 3.3%, P = 0.12).
Mutation Frequencies of Combined Mutation Types in Different Hepatitis B Virus Genotypes (%)
| Combined Mutation Types | Children | Adults | |||||
|---|---|---|---|---|---|---|---|
| Genotype B (n = 191) | Genotype C (n = 54) | P1 Value | Genotype B (n = 66) | Genotype C (n = 26) | P2 Value | P3 Value | |
| G1721A/G1757A | 0 | 14.8 | < 0.001 | 0 | 3.9 | 0.283 | 0.282 |
| G1721A/A1775G | 0 | 29.6 | < 0.001 | 0 | 3.9 | 0.283 | 0.008 |
| G1721A/C1856T | 0 | 16.7 | < 0.001 | 0 | 0 | - | 0.067 |
| G1721A/T1858C | 0 | 31.5 | < 0.001 | 0 | 3.9 | 0.283 | 0.006 |
| G1757A/A1775G | 0 | 14.8 | < 0.001 | 0 | 3.9 | 0.283 | 0.282 |
| G1757A/T1858C | 0 | 14.8 | < 0.001 | 0 | 3.9 | 0.283 | 0.282 |
| A1775G/C1856T | 0 | 14.8 | < 0.001 | 0 | 0 | - | 0.095 |
| A1775G/T1858C | 0 | 29.6 | < 0.001 | 0 | 3.9 | 0.283 | 0.008 |
| C1856T/T1858C | 0 | 16.7 | < 0.001 | 0 | 0.0 | - | 0.067 |
| G1757A/T1858C/G1721A | 0 | 14.8 | < 0.001 | 0 | 3.9 | 0.283 | 0.282 |
| G1757A/T1858C/A1775G | 0 | 14.8 | < 0.001 | 0 | 3.9 | 0.283 | 0.282 |
| G1757A/G1721A/A1775G | 0 | 14.8 | < 0.001 | 0 | 3.9 | 0.283 | 0.282 |
| T1858C/C1856T/G1721A | 0 | 16.7 | < 0.001 | 0 | 0 | - | 0.067 |
| T1858C/C1856T/A1775G | 0 | 14.8 | < 0.001 | 0 | 0.0 | - | 0.095 |
| T1858C/G1721A/A1775G | 0 | 29.6 | < 0.001 | 0 | 0 | - | 0.002 |
| C1856T/G1721A/A1775G | 0 | 14.8 | < 0.001 | 0 | 0.0 | - | 0.095 |
| A1775G/T1858C/C1856T/G1721A | 0 | 14.8 | < 0.001 | 0 | 0 | - | 0.095 |
| A1775G/T1858C/G1721A/G1757A | 0 | 14.8 | < 0.001 | 0 | 0 | - | 0.095 |
Correlations Analysis of Clinical Characteristics Between G1721A/A1775G/T1858C Triple-Mutation-Containing Children and Triple-Mutation-Free Group a
| Triple-Mutation-Containing Group (n = 16) | Triple-Mutation-Free Group (n = 229) | P Value | |
|---|---|---|---|
| Age, y | 4.56 ± 2.45 | 6.56 ± 4.12 | 0.012 |
| DNA load, log copies/mL | 7.56 ± 2.07 | 6.64 ± 2.29 | 0.045 |
| ALT, IU/L | 70.7 ± 62.2 | 51.5 ± 36.9 | 0.298 |
| HBeAg(+)/HBeAg(-) | 13/3 | 142/87 | 0.120 |
| Genotype, B/C | 0/16 | 191/38 | < 0.001 |
5. Discussion
Genotypes B and C are the predominant HBV genotypes in China (5). Infection with HBV genotype C strains compared with genotype B are associated with lower rates of HBeAg spontaneous clearance in serum, higher levels virus replication, more severe and advanced liver diseases, and decreased rate of response to IFN treatment (9). In this study, genotype B was the main genotype among children and adults in southern China (78.0% and 71.7%, respectively), followed by genotype C (22.0% and 28.3, respectively). The data of direct PCR sequencing showed that the mutation frequencies at 10 mutation positions in the BCP/precore regions, including nt1679, 1721, 1753, 1757, 1758, 1762, 1764, 1775, 1856 and 1858, were significantly higher in genotype C than in genotype B among children. In adults, only nt1679, 1758 and 1775 showed significantly higher mutation frequencies in HBV genotype C than in genotype B. The mutation ratio difference between genotypes B and C in children was higher than that of adults. Mutation frequencies of two attractive nucleotides, nt1896 and nt1899, did not show significant differences between patients genotypes B and C.
A previous study reported that T1858C mutations are often found in combination with A1775G (21). In our results, we showed that 16 of 17 T1858C mutation-containing sequences were in T1858C/A1775G/G1721A triple mutations. Statistical analysis demonstrated that the presence of a combined triple-mutation type, G1721A/A1775G/T1858C, was correlated with younger ages, genotype C strains, and a higher DNA load. In addition, other combined mutation types such as C1856T/T1858C/G1721A and G1721A/G1757A/A1775G/T1858C were also exclusively detected in children with genotype C infection, while the clinical characteristics of these combined mutation-containing patients showed no statistical difference compared to those without combined mutations. The molecular mechanism of these combined mutations with chronic HBV genotype C infection merits further investigations.
References
-
1.
Ott JJ, Stevens GA, Groeger J, Wiersma ST. Global epidemiology of hepatitis B virus infection: new estimates of age-specific HBsAg seroprevalence and endemicity. Vaccine. 2012;30(12):2212-9. [PubMed ID: 22273662]. https://doi.org/10.1016/j.vaccine.2011.12.116.
-
2.
Baumert TF, Thimme R, von Weizsacker F. Pathogenesis of hepatitis B virus infection. World J Gastroenterol. 2007;13(1):82-90. [PubMed ID: 17206757].
-
3.
Schaefer S. Hepatitis B virus taxonomy and hepatitis B virus genotypes. World J Gastroenterol. 2007;13(1):14-21. [PubMed ID: 17206751].
-
4.
Tatematsu K, Tanaka Y, Kurbanov F, Sugauchi F, Mano S, Maeshiro T, et al. A genetic variant of hepatitis B virus divergent from known human and ape genotypes isolated from a Japanese patient and provisionally assigned to new genotype J. J Virol. 2009;83(20):10538-47. [PubMed ID: 19640977]. https://doi.org/10.1128/JVI.00462-09.
-
5.
Hou J, Liu Z, Gu F. Epidemiology and Prevention of Hepatitis B Virus Infection. Int J Med Sci. 2005;2(1):50-7. [PubMed ID: 15968340].
-
6.
Orito E, Mizokami M, Sakugawa H, Michitaka K, Ishikawa K, Ichida T, et al. A case-control study for clinical and molecular biological differences between hepatitis B viruses of genotypes B and C. Japan HBV Genotype Research Group. Hepatology. 2001;33(1):218-23. [PubMed ID: 11124839]. https://doi.org/10.1053/jhep.2001.20532.
-
7.
Chu CJ, Hussain M, Lok AS. Hepatitis B virus genotype B is associated with earlier HBeAg seroconversion compared with hepatitis B virus genotype C. Gastroenterology. 2002;122(7):1756-62. [PubMed ID: 12055581].
-
8.
Ding X, Mizokami M, Yao G, Xu B, Orito E, Ueda R, et al. Hepatitis B virus genotype distribution among chronic hepatitis B virus carriers in Shanghai, China. Intervirology. 2001;44(1):43-7. [PubMed ID: 11223719].
-
9.
Kao JH, Chen PJ, Lai MY, Chen DS. Hepatitis B genotypes correlate with clinical outcomes in patients with chronic hepatitis B. Gastroenterology. 2000;118(3):554-9. [PubMed ID: 10702206].
-
10.
Kao JH, Wu NH, Chen PJ, Lai MY, Chen DS. Hepatitis B genotypes and the response to interferon therapy. J Hepatol. 2000;33(6):998-1002. [PubMed ID: 11131465].
-
11.
Chan HL, Tse CH, Mo F, Koh J, Wong VW, Wong GL, et al. High viral load and hepatitis B virus subgenotype ce are associated with increased risk of hepatocellular carcinoma. J Clin Oncol. 2008;26(2):177-82. [PubMed ID: 18182659]. https://doi.org/10.1200/JCO.2007.13.2043.
-
12.
Chan HL, Hui AY, Wong ML, Tse AM, Hung LC, Wong VW, et al. Genotype C hepatitis B virus infection is associated with an increased risk of hepatocellular carcinoma. Gut. 2004;53(10):1494-8. [PubMed ID: 15361502]. https://doi.org/10.1136/gut.2003.033324.
-
13.
Kay A, Zoulim F. Hepatitis B virus genetic variability and evolution. Virus Res. 2007;127(2):164-76. [PubMed ID: 17383765]. https://doi.org/10.1016/j.virusres.2007.02.021.
-
14.
Yang HI, Lu SN, Liaw YF, You SL, Sun CA, Wang LY, et al. Hepatitis B e antigen and the risk of hepatocellular carcinoma. N Engl J Med. 2002;347(3):168-74. [PubMed ID: 12124405]. https://doi.org/10.1056/NEJMoa013215.
-
15.
Chen CJ, Yang HI, Su J, Jen CL, You SL, Lu SN, et al. Risk of hepatocellular carcinoma across a biological gradient of serum hepatitis B virus DNA level. JAMA. 2006;295(1):65-73. [PubMed ID: 16391218]. https://doi.org/10.1001/jama.295.1.65.
-
16.
Wu CF, Yu MW, Lin CL, Liu CJ, Shih WL, Tsai KS, et al. Long-term tracking of hepatitis B viral load and the relationship with risk for hepatocellular carcinoma in men. Carcinogenesis. 2008;29(1):106-12. [PubMed ID: 17999990]. https://doi.org/10.1093/carcin/bgm252.
-
17.
Ghabeshi S, Sharifi Z, Hosseini SM, Mahmoodian Shooshtari M. Correlation between viral load of HBV in chronic hepatitis B patients and precore and Basal core promoter mutations. Hepat Mon. 2013;13(2). [PubMed ID: 23599717]. https://doi.org/10.5812/hepatmon.7415.
-
18.
Chauhan R, Kazim SN, Bhattacharjee J, Sakhuja P, Sarin SK. Basal core promoter, precore region mutations of HBV and their association with e antigen, genotype, and severity of liver disease in patients with chronic hepatitis B in India. J Med Virol. 2006;78(8):1047-54. [PubMed ID: 16789012]. https://doi.org/10.1002/jmv.20661.
-
19.
Lai MW, Lin TY, Tsao KC, Huang CG, Hsiao MJ, Liang KH, et al. Increased seroprevalence of HBV DNA with mutations in the s gene among individuals greater than 18 years old after complete vaccination. Gastroenterology. 2012;143(2):400-7. [PubMed ID: 22580098]. https://doi.org/10.1053/j.gastro.2012.05.002.
-
20.
Alexopoulou A, Karayiannis P. HBeAg negative variants and their role in the natural history of chronic hepatitis B virus infection. World J Gastroenterol. 2014;20(24):7644-52. [PubMed ID: 24976702]. https://doi.org/10.3748/wjg.v20.i24.7644.
-
21.
Nordin M, Ingman M, Lindqvist B, Kidd-Ljunggren K. Variability in the precore and core promoter region of the hepatitis B virus genome. J Med Virol. 2014;86(3):437-45. [PubMed ID: 24249691]. https://doi.org/10.1002/jmv.23839.