Abstract
Background:
Genome-wide association studies have shown that rs738491, rs2143571, and rs3761472 in the sorting and assembly machinery component 50 homolog (SAMM50) gene are significantly associated with susceptibility to nonalcoholic fatty liver disease (NAFLD).Objectives:
The present study evaluated the association between the three genetic variants in the SAMM50 gene and susceptibility to NAFLD in a Chinese Han population.Patients and Methods:
Genotypes for 3 single nucleotide polymorphisms (SNPs), viz rs738491, rs2143571, and rs3761472, in the SAMM50 gene were determined using an improved multiplex ligation detection reaction technique in 340 B-type ultrasonography-diagnosed NAFLD patients and 452 healthy controls. Meanwhile, serum lipid profiles and liver enzymes were estimated using standard clinical laboratory methods. The SNP-SNP interactions were analyzed by performing multifactor dimensionality reduction (MDR) and generalized multifactor dimensionality reduction (GMDR).Results:
The genotype and allele frequencies of the SAMM50 polymorphisms between the NAFLD group and the control group were significantly different (all Ps < 0.05). In the multivariate analysis adjusted for gender, age, and body mass index, the carriers of the rs738491 T allele, rs2143571 A allele, and rs3761472 G allele had significantly increased susceptibility to NAFLD (OR, 1.507; 95% CI, 1.035 to 2.195; P = 0.032; OR, 1.761; 95% CI, 1.232 to 2.517; P = 0.002; OR, 1.483; 95% CI, 1.039 to 2.115; P = 0.030, respectively). Moreover, the rs738491 T allele carriers had significantly higher levels of alanine aminotransferase (ALT) (P = 0.017) than did the noncarriers. However, differences in the levels of serum triglyceride (TG) and aspartate aminotransferase (AST) were not statistically significant (P = 0.123; P = 0.107). The Rs2143571 A allele and the rs3761472 G allele were both deeply associated with increased levels of serum TG, ALT, and AST (all Ps < 0.05). Furthermore, the MDR and GMDR showed that a synergistic relationship might exist between rs738491, rs2143571, and rs3761472 in the SAMM50 gene and the pathophysiology and genetics of NAFLD.Conclusions:
We first demonstrated that the rs738491 T allele, rs2143571 A allele, and rs3761472 G allele in the SAMM50 gene created susceptibility to NAFLD in a Chinese Han population. The combination of the three SNPs in the SAMM50 gene may have synergism to predict the predisposition to NAFLD.Keywords
Nonalcoholic Fatty Liver Disease Polymorphism Single Nucleotide SAMM50
1. Background
Nonalcoholic fatty liver disease (NAFLD) is the main cause of chronic liver disease and a major public health problem worldwide (1). Simple fatty liver is generally considered a benign pathological process. However, 20% of simple fatty liver patients display lobular inflammation and hepatocyte injury, a condition designated as “nonalcoholic steatohepatitis” (NASH), which may even progress toward liver cirrhosis and hepatocellular carcinoma (2, 3). The detailed pathogenesis of NAFLD remains unclear. Generally, it is believed that the genetic factor is important in the development of NAFLD (4-6), and specific ethnic groups or environmental conditions have been proven to influence the results. Identifying genetic associations with NAFLD in different ethnic groups will lead to the development of new noninvasive biomarkers for the early diagnosis of NAFLD and may allow early preventive and therapeutic strategies for people at high risk in different ethnic groups.
Recently, 3 single nucleotide polymorphisms (SNPs), viz rs738491, rs2143571, and rs3761472, in the sorting and assembly machinery component 50 homolog (SAMM50) gene were shown to be deeply associated with NAFLD after adjusting for age, gender, and body mass index (BMI) in the Japanese population (7). In Indian subjects, a subsequent study demonstrated a significant link between rs2143571 and NAFLD (8).
Now, NAFLD has been confirmed as the fastest growing etiology for hepatocellular carcinoma in China.
2. Objectives
Because the association between the 3 SNPs and NAFLD susceptibility in a Chinese Han population had not been investigated previously, the present study sought to explore the association between the genetic variants in rs738491, rs2143571, and rs3761472 in the SAMM50 gene and susceptibility to NAFLD in a Chinese Han population. The findings should, in turn, provide new insights into the pathophysiology and genetics of NAFLD.
3. Patients and Methods
3.1. Patients and controls
From September 2012 to October 2014, 340 unrelated Chinese NAFLD patients diagnosed by B-type ultrasonography (female, 189; male, 151; mean age, 41.54 ± 8.19 years) (9) and 452 healthy control subjects matched for sex and age (female, 228; male, 224; mean age, 40.18 ± 11.03 years) were enrolled in this study. All the subjects were recruited during the same study period from the Department of Gastroenterology and the Medical Center of Qingdao Municipal Hospital. The diagnosis of NAFLD was made according to the guidelines for the management of NAFLD of the Chinese Medical Association in 2010 (10). The exclusion criteria comprised alcohol abuse (≥ 210 g/week for males and ≥ 140 g/week for females), chronic viral hepatitis (hepatitis B and C), drug-induced steatosis, autoimmune liver disease, Wilson’s disease, and alpha-l-antitrypsin deficiency (5). The healthy controls were chosen from among persons without evidence of abnormal lipid profiles, abnormal liver enzymes, or any of the features of the metabolic syndrome (5).
The study was conducted according to the guidelines of the World Medical Association Declaration of Helsinki (11). All the subjects provided written informed consent in accordance with the study protocol approved by the Ethics Committee of Qingdao Municipal Hospital.
3.2. Demographic and Clinical Assessments
Demographic parameters such as BMI were determined in all the subjects. Venous blood samples from each participant were obtained and collected into sterile tubes containing ethylenediaminetetraacetic acid (EDTA). Serological assays, including serum concentrations of alanine aminotransferase (ALT), aspartate aminotransferase (AST), γ-glutamyltransferase (GGT), fasting plasma glucose (FPG), uric acid (UA), triglyceride (TG), total cholesterol (TC), high-density lipoprotein (HDL), and low-density lipoprotein (LDL), were carried out in accordance with the standard clinical laboratory methods in the Central Laboratory of Qingdao Municipal Hospital.
3.3. Genetic Analysis
Genomic DNA was isolated from noncoagulated blood samples using the Genomic DNA Purification Kit (BioTeke, Biotechnology, Beijing, China) following the user’s protocol. The genotyping of the 3 SNPs, viz rs738491, rs2143571, and rs3761472, in the SAMM50 gene was conducted using an improved multiplex ligation detection reaction technique (Genesky Biotechnologies Inc. Shanghai, China). Primers for rs738491, rs2143571, and rs3761472 were 5’-CTTCATTCCCCCTCCCATCATC-3’ (forward), 5’-ACAGGGGATGCAAGCCAAGAA-3’(reverse), 5’-GGAGTCCTGTCCTGCCACTTCTC-3’(forward), 5’-GGAAACCAGCAGTACCCACAGC-3’(reverse), 5’-CAATGGGAACCCAAAGTAAAATAACA-3’(forward), and 5’-GCCTGT AGCATACCATACTGCCTTCA-3’(reverse). The polymerase chain reaction (PCR) was performed as follows: denaturation at 95°C for 2 minutes, 11 cycles × (denaturation at 94°C for 20 seconds, annealing at 65°C - 0.5°C/cycle for 40 seconds, and elongation at 72°C for 1 minute 30 seconds), 24 cycles × (denaturation at 94°C for 20 seconds, annealing at 59°C for 30 seconds, and elongation at 72°C for 1 minute 30 seconds) at 72°C for 10 minutes, and keeping at 4°C. The ligation products were loaded into the ABI 3730XL, and the raw data were analyzed using GeneMapper 4.1 software (Applied Biosystems).
3.4. Statistical Analysis
The statistical analyses were performed using statistical package for the social sciences (SPSS), version 17.0 (SPSS Inc. Chicago, IL, USA). All the data are presented as Mean ± Standard Deviation (SD) for the continuous variables and as percentages for the categorical variables. The Hardy-Weinberg equilibrium was assessed using the χ2 test. Genotypes and allele frequencies were calculated using the χ2 test, and the distributions between the NAFLD patients and the healthy controls were analyzed using the Pearson χ2 test or the Fisher exact test. The strength of the association between the genetic variants and NAFLD was evaluated by logistic regression analysis and estimated by the odds ratio (OR) with a 95% confidence interval (CI). Differences in the baseline characteristics between the 2 groups were examined using the Student t-test, paired samples t-test, or the χ2 test. The SNP-SNP interactions were analyzed by open-source Java software multifactor dimensionality reduction (MDR), version 3.0.2 (12), and generalized multifactor dimensionality reduction (GMDR), version 0.9 (13). A P value < 0.05 was considered statistically significant.
4. Results
4.1. Demographic and Clinical Characteristics of the Nonalcoholic Fatty Liver Disease Patients and Healthy Controls
Table 1 shows the demographic and clinical characteristics of the NAFLD patients and the healthy controls. The 2 groups were matched for sex and age (both Ps > 0.05). The NAFLD patients had higher BMI, waist and hip circumferences, serum levels of ALT, AST, GGT, FPG, TG, TC, and LDL, and lower HDL than did the healthy controls (all Ps < 0.05).
Demographic and Clinical Characteristics of Nonalcoholic Fatty Liver Disease Patients and Healthy Controls a
| NAFLD Patients | Healthy Controls | χ2 or t | P Value | |
|---|---|---|---|---|
| Female, % | 151 (44.4) | 224 (49.6) | χ2 = 2.061 | 0.151 |
| Age, y | 41.54 ± 8.19 | 40.18 ± 11.03 | t = 1.909 | 0.057 |
| BMI, kg/m2 | 26.17 ± 2.60 | 23.18 ± 3.07 | t = 14.796 | < 0.001 |
| ALT, U/L | 42.25 ± 25.36 | 29.73 ± 15.54 | t = 8.046 | < 0.001 |
| AST, U/L | 25.86 ± 12.65 | 22.82 ± 8.81 | t = 3.784 | < 0.001 |
| GGT, U/L | 29.73 ± 25.48 | 18.35 ± 22.90 | t = 6.500 | < 0.001 |
| FPG, mmol/L | 5.51 ± 1.61 | 4.89 ± 0.92 | t = 6.418 | < 0.001 |
| UA, mmol/L | 360.54 ± 95.20 | 306.28 ± 82.12 | t = 8.400 | < 0.001 |
| TG, mmol/L | 2.40 ± 2.32 | 1.60 ± 1.97 | t = 5.175 | < 0.001 |
| TC, mmol/L | 5.00 ± 1.05 | 4.58 ± 1.01 | t = 5.574 | < 0.001 |
| HDL, mmol/L | 1.29 ± 0.46 | 1.47 ± 0.31 | t = -6.479 | < 0.001 |
| LDL, mmol/L | 3.21 ± 1.04 | 2.72 ± 0.83 | t = 7.116 | < 0.001 |
4.2. Genotypes and Allele Frequencies of rs738491, rs2143571, and rs3761472
The genotypes of each SNP were in the Hardy-Weinberg equilibrium in the two groups (PNAFLD = 0.112, 0.077, 0.071; Pcontrol = 0.879, 0.709, 0.767). As is described in Table 2, there were significant differences in the genotype and allele frequencies of the SAMM50 polymorphisms between the NAFLD group and the control group (all Ps < 0.05). Moreover, Table 3 demonstrates strong links between the 3 genetic variants in the SAMM50 gene and NAFLD. The carriers of the rs738491 T allele, rs2143571 A allele, and rs3761472 G allele had significantly increased susceptibility to NAFLD (OR, 1.712; 95% CI, 1.223 to 2.398; P = 0.002; OR, 1.940; 95% CI, 1.410 to 2.671; P < 0.001; OR, 1.758; 95% CI, 1.288 to 2.399; P < 0.001, respectively). After adjusting for confounders (i.e. gender, age, and BMI), we observed that the significant associations still existed (OR, 1.507; 95% CI, 1.035 to 2.195; P = 0.032; OR, 1.761; 95% CI, 1.232 to 2.517; P = 0.002; OR, 1.483; 95% CI, 1.039 to 2.115; P = 0.030, respectively).
Distribution of Genotypes and Allele Frequencies of Genetic Variants in SAMM50 in Subjects
| Genotypes and Allele | Nonalcoholic Fatty Liver Disease Patients a | Controls a | χ2 | P Value |
|---|---|---|---|---|
| rs738491 | ||||
| Genotypes | 25.467 | < 0.001 | ||
| CC | 66 (19.41) | 132 (29.20) | ||
| CT | 151 (44.41) | 226 (50.00) | ||
| TT | 123 (36.18) | 94 (20.80) | ||
| Alleles | 24.604 | < 0.001 | ||
| C | 283 (41.62) | 490 (54.20) | ||
| T | 397 (48.38) | 414 (45.80) | ||
| rs2143571 | ||||
| Genotypes | 33.300 | < 0.001 | ||
| GG | 76 (22.35) | 162 (35.84) | ||
| GA | 152 (44.71) | 214 (47.35) | ||
| AA | 112 (32.94) | 76 (16.81) | ||
| Alleles | 34.172 | < 0.001 | ||
| G | 304 (44.71) | 538 (59.51) | ||
| A | 376 (55.29) | 366 (40.49) | ||
| rs3761472 | ||||
| Genotype | 26.380 | < 0.001 | ||
| AA | 85 (25.00) | 167 (36.95) | ||
| GA | 153 (45.00) | 213 (47.12) | ||
| GG | 102 (30.00) | 72 (15.93) | ||
| Alleles | 26.527 | < 0.001 | ||
| A | 323 (47.50) | 547 (60.51) | ||
| G | 357 (52.50) | 357 (39.49) | ||
Odds Ratios for Nonalcoholic Fatty Liver Disease According to Genotypes of SAMM50 Single Nucleotide Polymorphisms in Study Group
| Unadjusted OR, 95% CI | P Value | Adjusted OR, 95%CI a | P Value | |
|---|---|---|---|---|
| rs738491 | ||||
| CC | 1 | 1 | ||
| CT + TT | 1.712 (1.223 - 2.398) | 0.002 | 1.507 (1.035 - 2.195) | 0.032 |
| rs2143571 | ||||
| GG | 1 | 1 | ||
| GA + AA | 1.940 (1.410 ~ 2.671) | < 0.001 | 1.761 (1.232 - 2.517) | 0.002 |
| rs3761472 | ||||
| AA | 1 | 1 | ||
| AG + GG | 1.758 (1.288 - 2.399) | < 0.001 | 1.483 (1.039 - 2.115) | 0.030 |
4.3. Association Between the 3 Genetic Variants in the SAMM50 Gene and Clinical Features
As is shown in Table 4, compared with the noncarriers, the carriers of the rs738491 T allele had significantly high levels of liver enzymes (serum ALT; P = 0.017) and lipid profiles (TC and LDL; both Ps < 0.05). Nevertheless, differences in the levels of serum AST, TG, and HDL between the carriers and the noncarriers of the rs738491 T allele were not statistically significant (P = 0.107; P = 0.123; P = 0.099). Similar strong links, except for the AST and TG levels of serum, were observed in the other 2 variants. The rs2143571 A allele and rs3761472 G allele were also both deeply associated with increased levels of serum AST and TG (P = 0.045; P = 0.002; P = 0.008; P = 0.040).
Association Between SAMM50 Polymorphisms and Clinical Features in Study Subjects a
| rs738491 | rs2143571 | rs3761472 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Noncarriers | Carriers | χ2 or t | P Value | Noncarriers | Carriers | χ2 or t | P Value | Noncarriers | Carriers | χ2 or t | P Value | |
| n | 198 | 594 | 238 | 554 | 252 | 540 | ||||||
| Female, % | 86 (43.4) | 289 (48.7) | χ2 = 1.622 | 0.203 | 114 (47.9) | 261 (47.1) | χ2 = 0.041 | 0.839 | 128 (50.8) | 247 (45.7) | χ2 = 1.760 | 0.185 |
| Age, y | 41.51 ± 10.08 | 40.51 ± 9.88 | T = 1.223 | 0.222 | 40.81 ± 10.37 | 40.74 ± 9.75 | t = 0.090 | 0.929 | 41.61 ± 10.78 | 40.37 ± 9.50 | t = 1.563 | 0.119 |
| BM, kg/m2 | 23.91 ± 2.94 | 24.65 ± 3.31 | t = -2.938 | 0.004 | 24.08 ± 3.24 | 24.63 ± 3.23 | t = -2.192 | 0.029 | 23.93±3.26 | 24.71±3.20 | t =-3.210 | 0.001 |
| ALT, U/L | 32.20 ± 18.82 | 36.08 ± 21.94 | t =-2.407 | 0.017 | 31.29 ± 18.63 | 36.75 ± 22.10 | t = -3.572 | < 0.001 | 30.95 ± 18.27 | 37.05 ± 22.26 | t = -4.069 | < 0.001 |
| AST, U/L | 23.06 ± 9.15 | 24.48 ± 11.19 | t = -1.613 | 0.107 | 23.09 ± 8.49 | 24.57 ± 11.54 | t = -2.012 | 0.045 | 22.63 ± 8.50 | 24.82 ± 11.56 | t = -2.680 | 0.008 |
| GGT, U/L | 20.62 ± 20.84 | 24.11 ± 25.78 | t = -1.725 | 0.085 | 19.37 ± 15.15 | 24.89 ± 27.63 | t = -3.605 | < 0.001 | 18.55 ± 15.79 | 25.42 ± 27.62 | t = -4.435 | < 0.001 |
| FPG, mmol/L | 4.95 ± 0.95 | 5.22 ± 1.39 | t = -3.075 | 0.002 | 5.07 ± 1.11 | 5.19 ± 1.37 | t = -1.399 | 0.162 | 5.06 ± 1.20 | 5.20 ± 1.34 | t = -1.501 | 0.134 |
| UA, mmol/L | 320.87 ± 95.35 | 332.36 ± 90.65 | t = -1.521 | 0.129 | 332.03 ± 100.30 | 328.41 ± 88.13 | t = 0.482 | 0.63 | 325.45 ± 90.31 | 331.38 ± 92.67 | t = -0.845 | 0.398 |
| TG, mmol/L | 1.76±1.84 | 2.00±2.25 | t =- 1.546 | 0.123 | 1.62±1.66 | 2.08±2.32 | t = -3.145 | 0.002 | 1.71±2.08 | 2.05±2.19 | t =- 2.055 | < 0.001 |
| TC, mmol/L | 4.41 ± 0.89 | 4.88 ±1.08 | t = -6.117 | < 0.001 | 4.57 ± 0.98 | 4.84 ± 1.07 | t = -3.306 | 0.001 | 4.74 ± 0.94 | 4.77 ± 1.10 | t = -0.436 | 0.663 |
| HDL, mmol/L | 1.44 ± 0.52 | 1.38 ± 0.34 | t = 1.651 | 0.099 | 1.43 ± 0.51 | 1.38 ± 0.33 | t = 1.456 | 0.146 | 1.41 ± 0.33 | 1.39 ± 0.42 | t = 0.587 | 0.557 |
| LDL, mmol/L | 2.69 ± 0.89 | 3.01 ± 0.96 | t =-4.129 | < 0.001 | 2.74 ± 0.83 | 3.01 ± 0.99 | t = -4.042 | < 0.001 | 2.61 ± 0.79 | 3.08 ± 0.99 | t = -6.691 | < 0.001 |
4.4. Analysis of SNP-SNP Interactions
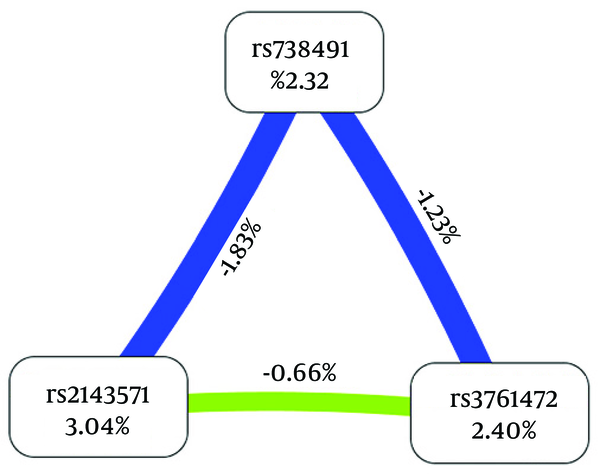
As is shown in Figures 1 and 2, the dendrogram and the Fruchterman-Reingold described the interactions between these SNPs. The results of the GMDR model analysis of the SNP-SNP interactions are demonstrated in Table 5. The results showed that rs2143571 was the best single-locus model to predict NAFLD (testing accuracy [TA], 54.94%; P = 0.0547; cross-validation consistency [CVC], 8/10). The best two-locus model was the combination of rs2143571 and rs3761472 (TA, 62.21%; P = 0.001; CVC, 10/10). The three-locus model included rs738491, rs2143571, and rs3761472 (TA, 60.79%; P = 0.001; CVC, 10/10). Therefore, the best model was the two-locus model, a combination of rs2143571 and rs3761472, with the highest TA and perfect CVC (OR, 2.9430; 95%CI, 1.9291 to 4.4899; P < 0.0001).
SNP-SNP Interaction Dendrogram
Fruchterman-Reingold, Describing Interactions Between the 3 Genetic Variants
Best Models to Predict Nonalcoholic Fatty Liver Disease by Generalized Multifactor Dimensionality Reduction (GMDR) a
5. Discussion
This study aimed to identify NAFLD susceptibility of the genetic variants in rs738491, rs2143571, and rs3761472 in the SAMM50 gene in a Chinese Han population. The GMDR analysis was employed to determine the SNP-SNP interactions.
Previous studies have indicated that the rs738491 T allele, rs2143571 A allele, and rs3761472 G allele in the SAMM50 gene may contribute to the progression from simple fatty liver to NASH, not only significantly associated with predisposition to NAFLD (7, 8), but also with decreased levels of serum TG and increased levels of ferritin and hyaluronic acid, which are high in NASH. However, our data demonstrated a controversial association inasmuch as the rs2143571 A allele and the rs3761472 G allele were both deeply associated with increased serum TG levels, whereas the rs738491 T allele did not show a significant P value. Furthermore, the ALT and AST levels of serum were significantly different between the carriers and the noncarriers of the rs2143571 A allele and the rs3761472 G allele, while the rs738491 T allele was only associated with increased levels of serum ALT. It is worthy of note that a similar finding has been confirmed in Indian subjects (8). This suggested that the genetic variants in rs2143571 and rs3761472, but not in rs738491, were associated with hepatic fat accumulation, as well as liver injury (13). Furthermore, adjusting for gender, age, and BMI, we demonstrated that the carriers of the rs738491 T allele, rs2143571 A allele, and rs3761472 G allele increased the predisposition to NAFLD compared with the noncarriers. However, the current results should be confirmed by future studies including larger and multiple ethnic populations.
The Sam50, encoded by the SAMM50 gene, is a part of the sorting and assembly machinery (SAM) required for the assembly pathway of the mitochondrial outer membrane β-barrel proteins (14). It is crucial for maintaining mitochondrial shape, morphology of mitochondrial cristae, and assembly of respiratory chain complexes (14, 15). Ott et al. confirmed that a moderate reduction in the Sam50 could result in changes in mitochondrial shape and the morphology of cristae and that depletion of the Sam50 could influence mitochondrial respiratory complexes (14). Various studies have established that mitochondrial dysfunction (loss of mitochondrial cristae and paracrystalline inclusions) plays an important role in insulin resistance (16, 17). Hepatic insulin resistance is known as the major pathophysiological contributor to the development and progression of NAFLD (18). In addition, mitochondrial abnormalities have been observed in the liver biopsy specimens of patients with NASH (19). These findings and our data indicate that the rs738491 T allele, rs2143571 A allele, and rs3761472 G allele in the SAMM50 gene may be involved in mitochondrial dysfunction and subsequent insulin resistance, resulting in the development and progression of NAFLD.
Given that NAFLD represents a complex disorder influenced by the interplay between genetic and environmental factors, multi-gene or SNP-SNP interaction studies may help discover the risk factors for NAFLD. Accordingly, we performed the MDR (12) and GMDR (20) to determine the potential SNP-SNP interactions between the 3 SNPs in the SAMM50 gene. The analysis of the SNP-SNP interactions showed a strong interaction between rs738491, rs2143571, and rs3761472 regarding susceptibility to NAFLD. As is described in Table 5, the results of the GMDR indicated that a synergistic relationship might exist between rs738491, rs2143571, and rs3761472 in the SAMM50 gene and the pathophysiology and genetic background of NAFLD. Additionally, a combination of rs2143571 and rs3761472 was the best model to predict the susceptibility to NAFLD compared to the single SNP alone.
However, several limitations existed in our study. First, the lack of liver biopsy is the main limitation of the present study. Liver biopsy is recognized as the gold standard in providing an accurate diagnosis of NAFLD; it is, however, expensive and not ethically feasible for uninvestigated subjects in epidemiological studies. We determined the hepatic fat content employing liver ultrasonography, which is widely utilized for the measurement of fatty liver disease (9) but cannot provide reliable quantitative information (21, 22). Moreover, liver ultrasonography cannot detect small changes in hepatic fat content over time, and its sensitivity and specificity are decreased in diagnosing obese NAFLD patients (23). Second, the underlying molecular mechanisms of the association and the SNP-SNP interactions should be explored by future basic and clinical studies.
We first demonstrated that genetic variants in rs738491, rs2143571, and rs3761472 in the SAMM50 gene created susceptibility to NAFLD in a Chinese Han population. The SNP-SNP interaction studies showed that a combination of the 3 SNPs in the SAMM50 gene might have synergism to predict the predisposition to NAFLD. The present study will help identify high-risk populations and perform appropriate interventions. Meanwhile, it will provide new ideas for the prevention and treatment of NAFLD.
References
-
1.
Loomba R, Sanyal AJ. The global NAFLD epidemic. Nat Rev Gastroenterol Hepatol. 2013;10(11):686-90. [PubMed ID: 24042449]. https://doi.org/10.1038/nrgastro.2013.171.
-
2.
Angulo P. Long-term mortality in nonalcoholic fatty liver disease: is liver histology of any prognostic significance? Hepatology. 2010;51(2):373-5. [PubMed ID: 20101746]. https://doi.org/10.1002/hep.23521.
-
3.
Review T, LaBrecque DR, Abbas Z, Anania F, Ferenci P, Khan AG, et al. World Gastroenterology Organisation global guidelines: Nonalcoholic fatty liver disease and nonalcoholic steatohepatitis. J Clin Gastroenterol. 2014;48(6):467-73. [PubMed ID: 24921212]. https://doi.org/10.1097/MCG.0000000000000116.
-
4.
Xu R, Tao A, Zhang S, Deng Y, Chen G. Association between patatin-like phospholipase domain containing 3 gene (PNPLA3) polymorphisms and nonalcoholic fatty liver disease: a HuGE review and meta-analysis. Sci Rep. 2015;5:9284. [PubMed ID: 25791171]. https://doi.org/10.1038/srep09284.
-
5.
Sookoian S, Castano GO, Scian R, Mallardi P, Fernandez Gianotti T, Burgueno AL, et al. Genetic variation in transmembrane 6 superfamily member 2 and the risk of nonalcoholic fatty liver disease and histological disease severity. Hepatology. 2015;61(2):515-25. [PubMed ID: 25302781]. https://doi.org/10.1002/hep.27556.
-
6.
Wang X, Liu Z, Peng Z, Liu W. The TM6SF2 rs58542926 T allele is significantly associated with non-alcoholic fatty liver disease in Chinese. J Hepatol. 2015;62(6):1438-9. [PubMed ID: 25687425]. https://doi.org/10.1016/j.jhep.2015.01.040.
-
7.
Kitamoto T, Kitamoto A, Yoneda M, Hyogo H, Ochi H, Nakamura T, et al. Genome-wide scan revealed that polymorphisms in the PNPLA3, SAMM50, and PARVB genes are associated with development and progression of nonalcoholic fatty liver disease in Japan. Hum Genet. 2013;132(7):783-92. [PubMed ID: 23535911]. https://doi.org/10.1007/s00439-013-1294-3.
-
8.
Kanth VV, Sasikala M, Rao PN, Steffie Avanthi U, Rao KR, Nageshwar Reddy D. Pooled genetic analysis in ultrasound measured non-alcoholic fatty liver disease in Indian subjects: A pilot study. World J Hepatol. 2014;6(6):435-42. [PubMed ID: 25018854]. https://doi.org/10.4254/wjh.v6.i6.435.
-
9.
Razavizade M, Jamali R, Arj A, Talari H. Serum parameters predict the severity of ultrasonographic findings in non-alcoholic fatty liver disease. Hepatobiliary Pancreat Dis Int. 2012;11(5):513-20. [PubMed ID: 23060397].
-
10.
Guidelines for management of nonalcoholic fatty liver disease: an updated and revised edition [in Chinese]. Chin J Hepatol. 2010;18(3):163-6.
-
11.
World Medical A. World Medical Association Declaration of Helsinki: ethical principles for medical research involving human subjects. JAMA. 2013;310(20):2191-4. [PubMed ID: 24141714]. https://doi.org/10.1001/jama.2013.281053.
-
12.
Moore JH, Gilbert JC, Tsai CT, Chiang FT, Holden T, Barney N, et al. A flexible computational framework for detecting, characterizing, and interpreting statistical patterns of epistasis in genetic studies of human disease susceptibility. J Theor Biol. 2006;241(2):252-61. [PubMed ID: 16457852]. https://doi.org/10.1016/j.jtbi.2005.11.036.
-
13.
Chen LZ, Xin YN, Geng N, Jiang M, Zhang DD, Xuan SY. PNPLA3 I148M variant in nonalcoholic fatty liver disease: demographic and ethnic characteristics and the role of the variant in nonalcoholic fatty liver fibrosis. World J Gastroenterol. 2015;21(3):794-802. [PubMed ID: 25624712]. https://doi.org/10.3748/wjg.v21.i3.794.
-
14.
Ott C, Ross K, Straub S, Thiede B, Gotz M, Goosmann C, et al. Sam50 functions in mitochondrial intermembrane space bridging and biogenesis of respiratory complexes. Mol Cell Biol. 2012;32(6):1173-88. [PubMed ID: 22252321]. https://doi.org/10.1128/MCB.06388-11.
-
15.
Ott C, Dorsch E, Fraunholz M, Straub S, Kozjak-Pavlovic V. Detailed analysis of the human mitochondrial contact site complex indicate a hierarchy of subunits. PLoS One. 2015;10(3):e0120213. [PubMed ID: 25781180]. https://doi.org/10.1371/journal.pone.0120213.
-
16.
Lowell BB, Shulman GI. Mitochondrial dysfunction and type 2 diabetes. Science. 2005;307(5708):384-7. [PubMed ID: 15662004]. https://doi.org/10.1126/science.1104343.
-
17.
Ma ZA, Zhao Z, Turk J. Mitochondrial dysfunction and beta-cell failure in type 2 diabetes mellitus. Exp Diabetes Res. 2012;2012:703538. [PubMed ID: 22110477]. https://doi.org/10.1155/2012/703538.
-
18.
Rector RS, Thyfault JP, Uptergrove GM, Morris EM, Naples SP, Borengasser SJ, et al. Mitochondrial dysfunction precedes insulin resistance and hepatic steatosis and contributes to the natural history of non-alcoholic fatty liver disease in an obese rodent model. J Hepatol. 2010;52(5):727-36. [PubMed ID: 20347174]. https://doi.org/10.1016/j.jhep.2009.11.030.
-
19.
Sanyal AJ, Campbell-Sargent C, Mirshahi F, Rizzo WB, Contos MJ, Sterling RK, et al. Nonalcoholic steatohepatitis: association of insulin resistance and mitochondrial abnormalities. Gastroenterology. 2001;120(5):1183-92. [PubMed ID: 11266382]. https://doi.org/10.1053/gast.2001.23256.
-
20.
Lou XY, Chen GB, Yan L, Ma JZ, Zhu J, Elston RC, et al. A generalized combinatorial approach for detecting gene-by-gene and gene-by-environment interactions with application to nicotine dependence. Am J Hum Genet. 2007;80(6):1125-37. [PubMed ID: 17503330]. https://doi.org/10.1086/518312.
-
21.
Saadeh S, Younossi ZM, Remer EM, Gramlich T, Ong JP, Hurley M, et al. The utility of radiological imaging in nonalcoholic fatty liver disease. Gastroenterology. 2002;123(3):745-50. [PubMed ID: 12198701].
-
22.
Charatcharoenwitthaya P, Lindor KD. Role of radiologic modalities in the management of non-alcoholic steatohepatitis. Clin Liver Dis. 2007;11(1):37-54. [PubMed ID: 17544971]. https://doi.org/10.1016/j.cld.2007.02.014.
-
23.
Mottin CC, Moretto M, Padoin AV, Swarowsky AM, Toneto MG, Glock L, et al. The role of ultrasound in the diagnosis of hepatic steatosis in morbidly obese patients. Obes Surg. 2004;14(5):635-7. [PubMed ID: 15186630]. https://doi.org/10.1381/096089204323093408.