Abstract
Background:
The functional and prognostic significance of Ras association domain family 1A gene (RASSF1A) on hepatocellular carcinoma (HCC) has not been well characterized.Objectives:
This study aimed to investigate the association between Ala133Ser polymorphism or promoter methylation in RASSF1A and the prognosis of HCC in Nantong City, one of the areas with the highest incidence of cancer in China.Patients and Methods:
Using peripheral blood plasma, the incidence rate of RASSF1A Ala133Ser in 235 controls and subjects with 260 HCC was analyzed by the polymerase chain reaction and sequencing. We further investigated the RASSF1A methylation status in HCC and corresponding peri-tumorous normal tissues using the methylation-specific polymerase chain reaction approach.Results:
It was found that the frequency of the RASSF1A Ala133Ser T allele (Ala/Ser and Ser/Ser) genotype in HCC cases was observably higher than that of normal subjects (P < 0.001). In comparison to the Ala/Ala genotype, the T allele genotype improved the susceptibility to HCC. The study also found that RASSF1A methylation improves the risk of HCC. Furthermore, in contrast with the corresponding peri-tumorous normal tissues, we observed that the RASSF1A methylation status was markedly higher in HCC tissues (P < 0.001). The Kaplan-Meier and multivariate analyses suggested that the poor survival of HCC patients was closely connected with hepatocirrhosis, Barcelona Clinic Liver Cancer stage, Edmondson division, RASSF1A methylation and Ala133Ser polymorphism (P < 0.001).Conclusions:
The polymorphism and promoter methylation of RASSF1A may be a significant factor in HCC, and can be an indicator for poor prognosis in patients with HCC.Keywords
Hepatocellular Carcinoma Polymorphism Methylation Prognosis Susceptibility
1. Background
The highest rates of primary liver cancer are found in East and South-East Asia. Hepatocellular carcinoma (HCC) is the primary histological type of hepatic carcinoma, accounting for 85 - 90% of the total burden of liver cancer all over the world (1, 2). The occurrence and death rate of HCC are almost the same as its high lethality rate (2). The onset of HCC has a multi-factorial and multi-stage course, involving both hereditary and environmental factors. The long-term carcinogenic effects give rise to genetic changes, which can lead to tumor formation (3). It is necessary to search for hereditary factors, which could help us detect the population at highest risk and better regulate the screening procedures. Besides, the recognition of risk factors could lead to better diagnoses and planning of novel prevention measures for high-risk individuals (4).
As an anti-oncogene, the Ras association domain family 1A gene (RASSF1A) has been reported to play a vital role in the maintenance of genomic instability; it controls a sequence of vital cellular functions in the integration of signaling pathways (5-9). The loss of expression by promoter methylation of RASSF1A is one of the most common early events in HCC that plays an important role in tumorigenesis and metastasis of HCC (10, 11). The most widespread form of human genetic variation is single nucleotide polymorphism (SNP), which may contribute to tumor sensitivity. To date, many studies have demonstrated that SNPs of RASSF1A are associated with the risk of many types of cancers, including breast cancer (12-15), lung cancer (16, 17), esophageal cancer (16, 18), head and neck cancer (16), colorectal cancer (16), renal cell carcinoma (19), HCC (20), gastric cancer (18), and prostate cancer (21). It has been proven that guanine (G)/thymine (T) SNP at the first position of codon 133 in exon 3 of RASSF1A (dbSNP ID: rs2073498), leads to the replacement of an Ala residue (GCT) through the Ser residue (TCT) in the ataxia telangiectasia mutated (ATM) phosphorylated position, and can impact the function of RASSF1A (22, 23). However, the results of these studies have been controversial (12-20). This is because the frequency of the Ser allele of RASSF1A Ala133Ser polymorphism varies in different geographic areas and ethnic populations (24).
The Qidong county of Nantong city is one of the regions with the highest incidence of HCC in China (25). In the recent years, several studies have investigated the correlation between genetic polymorphisms, methylation and HCC. Nevertheless, the effects of RASSF1A Ala133Ser polymorphism and methylation on HCC have not been well identified in Nantong.
Determining the molecular spectroscopy of HCC is essential as it could help us recognize molecular biomarkers for the screening of at-risk individuals and achieve early detection.
2. Objectives
A hospital-based case control research including 260 HCC cases and 235 controls was carried out to investigate the impact of RASSF1A Ala133Ser polymorphism and promoter methylation on HCC, and its prognostic and functional implications in subjects from a high incidence area in Nantong.
3. Patients and Methods
3.1. Study Population
The tumor and corresponding peri-tumorous normal tissues were obtained from 260 HCC patients whose frozen tumor samples had been collected from an affiliated hospital of Nantong University, from March 2007 to December 2012. In addition, 235 non-cancer subjects from the same hospital were treated as normal controls and matched according to their age, gender and ethnicity. We gathered 5 mL of pre-operation peripheral blood samples from each of the selected patients and control subjects in succession. Before the surgery, the patients were not treated by radiation, chemical or immunization therapy. The average age of the patients at the time of surgery was 53.6 years. Additional clinical data were recorded and are presented in Table 1. We confirmed the tumor stage in accordance with the barcelona clinic liver cancer (BCLC) staging system (26). Written informed consents and any related pictures were obtained from each patient prior to publication of this study. We received an ethics approval to do this research from the Human Research Ethics Committee of Nantong University affiliated hospital, Jiangsu province, China. Overall survival of these 260 patients was followed until July 31st, 2013. During the final follow-up, the data of surviving patients without symptoms of disease was examined.
The Genotyping of Ala133Ser in RASSF1A Gene in Patients With Hepatocellular Carcinoma and Normal Controls
| Genotyping | Control a | HCC a | P | OR | 95% CI |
|---|---|---|---|---|---|
| Allele frequency | |||||
| Ala | 445 (94.7) | 452 (86.9) | |||
| Ser | 25 (5.3) | 68 (13.1) | < 0.001 | 2.678 | 1.662 - 4.314 |
| General genotype | |||||
| Ala/Ala | 211 (89.8) | 200 (76.9) | |||
| Ala/Ser | 23 (9.8) | 52 (20.0) | 0.001 | 2.385 | 1.407 - 4.042 |
| Ser/Ser | 1 (0.4) | 8 (3.1) | 0.017 | 8.440 | 1.046 - 68.089 |
| Dominant genotype | |||||
| Ala/Ala | 211 (89.8) | 200 (76.9) | |||
| Ala/Ser + Ser/Ser | 24 (10.2) | 60 (23.1) | < 0.001 | 2.638 | 1.582 - 4.398 |
| Recessive genotype | |||||
| Ala/Ala + Ala/Ser | 234 (99.6) | 252 (96.9) | |||
| Ser/Ser | 1 (0.4) | 8 (3.1) | 0.027 | 7.429 | 0.922 - 59.847 |
3.2. Sequencing Analysis for Genotyping
Using the phenol/chloroform extraction method, genomic DNA was extracted from ambient blood plasma. The polymerase chain reaction (PCR) was carried out in a 50-μL reaction mixture, which contained 1 μL of genomic DNA, 10 mM dNTP, 10X buffered solution, 10 μM up and down treating agents (separately), and 1 U Taq DNA polymerase. The reagents for this study were purchased from TaKaRa in Beijing city, China. The sequences of the outside and inside primers were as follows: 5-’GCCAAATGATTCTGTCTTTCCCT forward and 5’-CAAGATAACCTCAGTTGTGACCCTC reverse. We used the GeneAmp RCR system 9700 gene magnifier (ABI, California, USA) to establish the conditions as follows: five minutes of pre-degeneration at 95°C; then 30 seconds of 30 cycles at 95°C, 30 seconds at 58°C and 30 seconds at 68°C; followed by 10 minutes of extension at 68°C. The purified PCR product was recovered according to the instructions in the PCR Purification Kit (Axygen, California, USA). After sequencing, the basic local alignment search tool (BLAST) was used to test and compare the results with Genbank.
3.3. Methylation-Specific Polymerase Chain Reaction (MSP)
Genomic DNA was isolated from frozen tissue specimens and cell lines by RNase treatment, phenol/chloroform extraction and ethanol precipitation. The bisulfite modifications of DNA and MSP were performed as described previously (27). Two cell lines, Hep3B (HCC cell line) and L02 (normal liver cell line), from the Chinese National Human Genome Center (Shanghai, China), were used as positive controls for methylated and un-methylated DNA. A ddH2O blank served as the negative control. Methylated samples were defined by the presence of methylated PCR products.
3.4. Statistical Analysis
The statistical comparisons were performed by the χ2 test using the SPSS 19.0 software (SPSS, Chicago, USA). The expected genotype frequencies measured by the Hardy-Weinberg equilibrium theory were used to compare the results with observed values. The odds ratios (ORs) and 95% confidence intervals (CIs) from the binary logistic regression analysis were used to estimate the associations between RASSF1A Ala133Ser genotypes and the risk of HCC. Survival curves were calculated by the Kaplan-Meier method. The factors of prognostic significance were successively investigated with the univariate and multivariate Cox regression model. For all tests, the significance level for statistical analysis was set at P < 0.05.
4. Results
4.1. Genotype Frequency Distribution of RASSF1A Ala133Ser Polymorphism in Hepatocellular Carcinoma
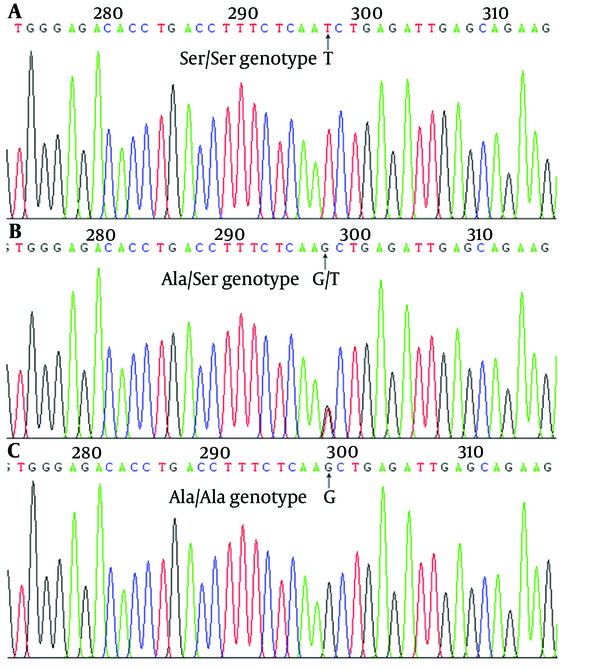
Peripheral blood plasmas taken from 235 controls and 260 HCC subjects were analyzed for the existence of Ala133Ser (Figure 1 and Table 1). The genotypic frequencies of the HCC patients (n = 260, χ2 = 3.760, P = 0.153) and controls (n = 235, χ2 = 0.188, P = 0.910) were both in the Hardy-Weinberg equilibrium. The results showed no population stratification and no sampling bias. There was a significant difference in the RASSF1A Ale133Ser T allele (Ala/Ser and Ser/Ser) genotype between the HCC patients and control subjects (23.1% vs. 10.2%, P < 0.001). With the Ala/Ala genotype as the reference, the Ala/Ser genotype increased the risk of HCC (OR = 2.385, 95% CI = 1.407 - 4.042, P = 0.001). Besides, in comparison with the Ala/Ala genotype, cases carrying the Ser/Ser genotype had an 8.440-fold increase in the risk of HCC (95% CI = 1.046 - 68.089, P = 0.017). Furthermore, Ser was found to have a significant gene dosage effect. With the Ala/Ala genotype as the reference, the OR for the T allele genotype was 2.638 (95% CI = 1.582 - 4.398, P < 0.001). Compared with the homozygote Ala/Ala and heterozygote Ala/Ser carriers, there was a significant increase in the risk of HCC in subjects with the homozygote variant of Ser/Ser of RASSF1A Ala133Ser polymorphism (OR = 7.429, 95% CI = 0.922 - 59.847, P = 0.027). The Ser frequency of RASSF1A Ala133Ser polymorphism in cases with HCC was significantly higher compared with the healthy control subjects (5.3% vs. 13.1%, P < 0.001). Furthermore, the Ala/Ala genotype in HCC peripheral blood plasma with hepatocirrhosis was more risky for patients with HCC than for healthy controls (OR = 10.189, 95% CI = 2.403 - 43.203, P < 0.001) (Table 2).
Sequencing Analysis of RASSF1A in Probands
The Correlation Between RASSF1A Ala133Ser Polymorphism and Suspected Risk Factorsa
| Parameters | Total | RASSF1A Ala133Ser Polymorphism b | P Value c | |
|---|---|---|---|---|
| Ala/Ala | Ala/Ser + Ser/Ser | |||
| Age, y | 0.147 | |||
| ≤ 55 | 126 | 92 (73.0) | 34 (27.0) | |
| > 55 | 134 | 108 (80.6) | 26 (19.4) | |
| Gender | 0.210 | |||
| Male | 144 | 115 (79.9) | 29 (20.1) | |
| Female | 116 | 85 (73.3) | 31 (26.7) | |
| Hepatocirrhosis | < 0.001 | |||
| Negative | 54 | 52 (96.3) | 2 (3.7) | |
| Positive | 206 | 148 (71.8) | 58 (28.2) | |
| AFP, μg/L | 0.867 | |||
| ≤ 20 | 54 | 42 (77.8) | 12 (22.2) | |
| > 20 | 206 | 158 (76.7) | 48 (23.3) | |
| Tumor size, cm | 0.288 | |||
| ≤ 3 | 167 | 125 (74.9) | 42 (25.1) | |
| > 3 | 93 | 75 (80.6) | 18 (19.4) | |
| BCLC stage | 0.559 | |||
| A | 198 | 154 (77.8) | 44 (22.2) | |
| B + C + D | 62 | 46 (74.2) | 16 (25.8) | |
| Edmondson division | 0.061 | |||
| G1 | 42 | 37 (88.1) | 5 (11.9) | |
| G2-4 | 218 | 163 (74.8) | 55 (25.2) | |
| HBV | 0.978 | |||
| Negative | 56 | 43 (76.8) | 13 (23.2) | |
| Positive | 204 | 157 (77.0) | 47 (23.0) | |
| Cigarette smoking | 0.064 | |||
| Non-smoker | 91 | 64 (70.3) | 27 (29.7) | |
| Smoker | 169 | 136 (80.5) | 33 (19.5) | |
| Alcohol consumption | 0.812 | |||
| Non-drinker | 90 | 70 (77.8) | 20 (22.2) | |
| Drinker | 170 | 130 (76.5) | 40 (23.5) | |
4.2. The Methylation of RASSF1A in Hepatocellular Carcinoma
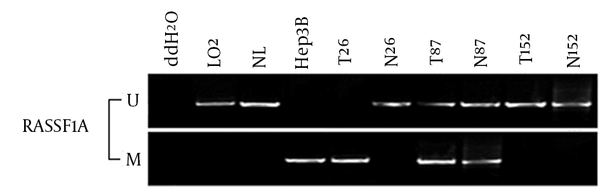
The frequency of RASSF1A methylation in HCC was 2.14 folds higher than the adjacent normal tissue (214/260, 82.3% vs. 101/260, 38.8%, P < 0.001). It also increased the risk of developing HCC by 7.324 folds (OR = 7.324, 95% CI = 4.887 - 10.975, P < 0.001) (Figure 2). Furthermore, the RASSF1A methylation ratio of G2-4 Edmondson division was significantly higher than that of G1 Edmondson division in HCC patients (OR = 2.165, 95% CI = 1.009 - 4.642, P = 0.044) (Table 3).
Methylation-Specific Polymerase Analysis of the RASSF1A Gene in Hepatocellular Carcinoma
Correlation Between RASSF1A Methylation and Suspected Risk Factorsa
| Parameters | Total | RASSF1A Methylation Status b | P Value c | |
|---|---|---|---|---|
| Methylated | Unmethylated | |||
| Age, y | 0.924 | |||
| ≤ 55 | 126 | 104 (82.5) | 22 (17.5) | |
| > 55 | 134 | 110 (82.1) | 24 (17.9) | |
| Gender | 0.619 | |||
| Male | 116 | 97 (83.6) | 19 (16.4) | |
| Female | 144 | 117 (81.2) | 27 (18.8) | |
| Hepatocirrhosis | 0.075 | |||
| Negative | 54 | 40 (74.1) | 14 (25.9) | |
| Positive | 206 | 174 (84.5) | 32 (15.5) | |
| AFP, μg/L | 0.306 | |||
| ≤ 20 | 54 | 47 (87.0) | 7 (13.0) | |
| > 20 | 206 | 167 (81.1) | 39 (18.9) | |
| Tumor size, cm | 0.123 | |||
| ≤ 3 | 167 | 142 (85.0) | 25 (15.0) | |
| > 3 | 93 | 72 (77.4) | 21 (22.6) | |
| BCLC stage | 0.439 | |||
| A | 198 | 165 (83.3) | 33 (16.7) | |
| B + C + D | 62 | 49 (79.0) | 13 (21.0) | |
| Edmondson division | 0.044 | |||
| G1 | 42 | 30 (71.4) | 12 (28.6) | |
| G2-4 | 218 | 184 (84.4) | 34 (15.6) | |
| HBV | 0.971 | |||
| Negative | 56 | 46 (82.1) | 10 (17.9) | |
| Positive | 204 | 168 (82.4) | 36 (17.6) | |
| Cigarette smoking | 0.082 | |||
| Non-smoker | 91 | 80 (87.9) | 11 (12.1) | |
| Smoker | 169 | 134 (79.3) | 35 (20.7) | |
| Alcohol consumption | 0.293 | |||
| Non-drinker | 90 | 71 (78.9) | 19 (21.1) | |
| Drinker | 170 | 143 (84.1) | 27 (15.9) | |
4.3. Survival Analysis
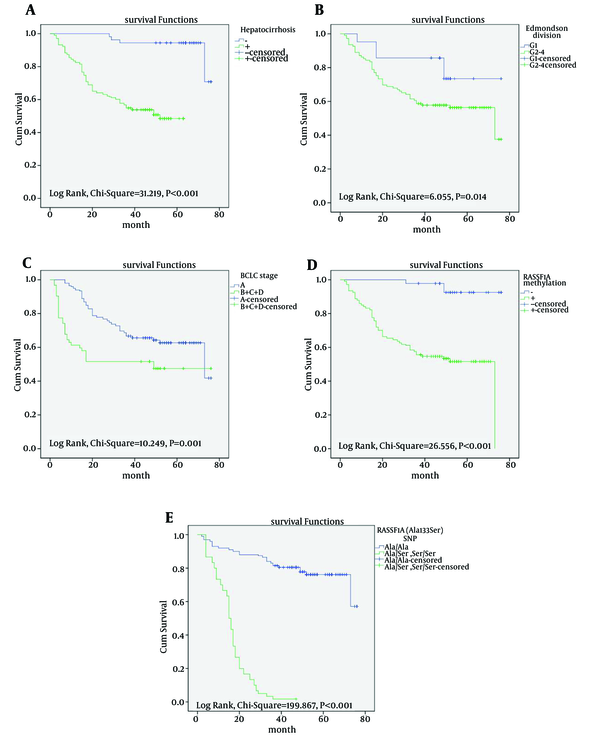
Univariate and multivariate Cox regression analyses verified that hepatocirrhosis, BCLC stage, Edmondson division, RASSF1A methylation and Ala133Ser polymorphism were the strongest prediction factors of overall survival (P < 0.05) (Table 4). The Kaplan-Meier survival curves revealed that there was a significantly better prognosis for patients with HCC carrying the Ala/Ala genotype without hepatocirrhosis and RASSF1A methylation, but with early BCLC stage, well-Edmondson division (Figure 3).
Univariate and Multivariable Analysis of Prognostic Factors for Overall Survival in Hepatocellular Carcinomaa
| Variable | Univariate Analysis | Multivariate Analysis | ||
|---|---|---|---|---|
| Hazard Ratio (95% CI) | P Value b | Hazard Ratio (95% CI) | P Value b | |
| Age, y | ||||
| ≤ 55 vs. > 55 | 0.668 (0.430 - 1.039) | 0.073 | ||
| Gender | ||||
| Male vs. female | 0.875 (0.579 - 1.324) | 0.528 | ||
| Cigarette smoking | ||||
| Non-smoker vs. smoker | 0.963 (0.622 - 1.491) | 0.866 | ||
| Alcohol consumption | ||||
| Non-drinker vs. drinker | 1.022 (0.648 - 1.612) | 0.925 | ||
| HBV | ||||
| Negative vs. Positive | 0.650 (0.371 - 1.138) | 0.132 | ||
| AFP, μg/L | ||||
| ≤ 20 vs. > 20 | 1.317 (0.792 - 2.190) | 0.288 | ||
| Tumor size, cm | ||||
| ≤ 3 vs. > 3 | 0.725 (0.445 - 1.181) | 0.196 | ||
| Hepatocirrhosis | ||||
| Negative vs. Positive | 9.035 (2.782 - 29.345) | < 0.001 | 7.728 (2.396 - 24.923) | 0.001 |
| BCLC stage | ||||
| A vs. B + C + D | 10.166 (5.359 - 19.285) | < 0.001 | 8.026 (4.693 - 13.725) | < 0.001 |
| Edmondson division | ||||
| G1 vs. G2-4 | 11.726 (5.036 - 27.302) | < 0.001 | 11.306 (5.132 - 24.907) | < 0.001 |
| RASSF1A methylation | ||||
| Negative vs. Positive | 7.904 (2.352 - 26.567) | 0.001 | 8.341 (2.531 - 27.491) | < 0.001 |
| RASSF1A (Ala133Ser) SNP | ||||
| Ala/Ala vs. Ala/Ser + Ser/Ser | 8.519 (5.127 - 14.157) | < 0.001 | 8.152i (5.045 - 13.173) | < 0.001 |
Kaplan-Meier Survival Analysis of Patients With Hepatocellular Carcinoma
5. Discussion
In this study, we investigated whether the RASSF1A Ala133Ser polymorphism could have an impact on the susceptibility to HCC. To the best of our knowledge, this is the first epidemiological study to reveal the association between RASSF1A Ala133Ser polymorphism and HCC risk in China. Our results suggest that the Ser allele of RASSF1A significantly increases the risk of HCC. The RASSF1A Ala133Ser polymorphism is located in a putative ATM phosphorylation site consensus sequence (7, 28). The cell cycle checkpoint responses at G1, S and G2 are all remarkably abnormal in ATM deficient cells (29). The Ser residue at the ATM site can be phosphorylated upon ionizing radiation and it can then induce cell death and suppress colony formation (30). Due to the involvement of ATM in DNA damage control, patients with HCC carrying RASSF1A Ala133Ser polymorphisms may be resistant to radiation-based chemotherapy (31). Therefore, it is reasonable to assume that subjects carrying the Ser allele of the RASSF1A Ala133Ser polymorphism may be susceptible to HCC. Knowledge about the mechanisms involved in HCC carcinogenesis may help identify targets for chemoprevention or therapeutic strategies.
The connection between RASSF1A Ala133Ser polymorphism and the risk of various cancers has been investigated by several studies (24). In this study, the frequency of genotypes containing the Ser allele of RASSF1A Ala133Ser polymorphism is similar to the other Asian cancer types, yet lower than in Caucasian cancer types. The difference in results may be explained by the following points. First, different types of cancer may have different mechanisms of carcinogenesis. Second, the discrepancy could also be interpreted partially by the influence of the gene-environment interaction in a multistep process of carcinogenesis. Third, studies recruiting controls form hospital populations are more likely to acquire significant results in allele comparison, heterozygous and dominant genetic models. Fourth, different cancer risks were also found in the studies using different genotyping methods. In this study, we also investigated promoter methylation of RASSF1A in HCC. Our results suggest that RASSF1A methylation may be an early event in HCC carcinogenesis. Further studies are required to characterize RASSF1A methylation in HCC precancerous lesions.
In order to confirm whether RASSF1A methylation and its Ala133Ser polymorphism are risk-factors predicting shorter survival after surgery, we also analyzed the influence of methylation and Ala133Ser polymorphism on patient survival. The Kaplan-Meier analysis indicated that the life span of patients with RASSF1A methylation was shorter in comparison to that of patients lacking this genetic feature. Besides, for the first time, we found that HCC patients with the Ser allele of RASSF1A Ala133Ser polymorphism had significantly poorer overall survival in Nantong. Univariate and further multivariate analyses demonstrated that hepatocirrhosis, BCLC stage, Edmondson division, RASSF1A methylation and RASSF1A Ala133Ser polymorphism independently predicted the unfavorable overall survival of patients with HCC.
In conclusion, this was the first study to reveal that the RASSF1A Ala133Ser polymorphism might confer genetic susceptibility to HCC in the Chinese population. The Ser allele of RASSF1A Ala133Ser and RASSF1A promoter methylation could significantly increase the risk of HCC development. Of course, further work is necessary to illustrate the mechanism of RASSF1A in the development of HCC. The RASSF1A gene will be developed ultimately as one of the molecular biomarkers for high-risk subject screening and early detection of HCC in the future.
Acknowledgements
References
-
1.
El-Serag HB, Rudolph KL. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132(7):2557-76. [PubMed ID: 17570226]. https://doi.org/10.1053/j.gastro.2007.04.061.
-
2.
Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61(2):69-90. [PubMed ID: 21296855]. https://doi.org/10.3322/caac.20107.
-
3.
Tanabe KK, Lemoine A, Finkelstein DM, Kawasaki H, Fujii T, Chung RT, et al. Epidermal growth factor gene functional polymorphism and the risk of hepatocellular carcinoma in patients with cirrhosis. JAMA. 2008;299(1):53-60. [PubMed ID: 18167406]. https://doi.org/10.1001/jama.2007.65.
-
4.
Llovet JM, Fuster J, Bruix J, Barcelona-Clinic Liver Cancer G. The Barcelona approach: diagnosis, staging, and treatment of hepatocellular carcinoma. Liver Transpl. 2004;10(2 Suppl 1):S115-20. [PubMed ID: 14762851]. https://doi.org/10.1002/lt.20034.
-
5.
Gordon M, Baksh S. RASSF1A: Not a prototypical Ras effector. Small GTPases. 2011;2(3):148-57. [PubMed ID: 21776416]. https://doi.org/10.4161/sgtp.2.3.16286.
-
6.
Donninger H, Vos MD, Clark GJ. The RASSF1A tumor suppressor. J Cell Sci. 2007;120(Pt 18):3163-72. [PubMed ID: 17878233]. https://doi.org/10.1242/jcs.010389.
-
7.
van der Weyden L, Adams DJ. The Ras-association domain family (RASSF) members and their role in human tumourigenesis. Biochim Biophys Acta. 2007;1776(1):58-85. [PubMed ID: 17692468]. https://doi.org/10.1016/j.bbcan.2007.06.003.
-
8.
Richter AM, Pfeifer GP, Dammann RH. The RASSF proteins in cancer; from epigenetic silencing to functional characterization. Biochim Biophys Acta. 2009;1796(2):114-28. [PubMed ID: 19344752]. https://doi.org/10.1016/j.bbcan.2009.03.004.
-
9.
Ortiz-Vega S, Khokhlatchev A, Nedwidek M, Zhang XF, Dammann R, Pfeifer GP, et al. The putative tumor suppressor RASSF1A homodimerizes and heterodimerizes with the Ras-GTP binding protein Nore1. Oncogene. 2002;21(9):1381-90. [PubMed ID: 11857081]. https://doi.org/10.1038/sj.onc.1205192.
-
10.
Hu L, Chen G, Yu H, Qiu X. Clinicopathological significance of RASSF1A reduced expression and hypermethylation in hepatocellular carcinoma. Hepatol Int. 2010;4(1):423-32. [PubMed ID: 20305761]. https://doi.org/10.1007/s12072-010-9164-8.
-
11.
Schagdarsurengin U, Wilkens L, Steinemann D, Flemming P, Kreipe HH, Pfeifer GP, et al. Frequent epigenetic inactivation of the RASSF1A gene in hepatocellular carcinoma. Oncogene. 2003;22(12):1866-71. [PubMed ID: 12660822]. https://doi.org/10.1038/sj.onc.1206338.
-
12.
Schagdarsurengin U, Seidel C, Ulbrich EJ, Kolbl H, Dittmer J, Dammann R. A polymorphism at codon 133 of the tumor suppressor RASSF1A is associated with tumorous alteration of the breast. Int J Oncol. 2005;27(1):185-91. [PubMed ID: 15942659].
-
13.
Gao B, Xie XJ, Huang C, Shames DS, Chen TT, Lewis CM, et al. RASSF1A polymorphism A133S is associated with early onset breast cancer in BRCA1/2 mutation carriers. Cancer Res. 2008;68(1):22-5. [PubMed ID: 18172292]. https://doi.org/10.1158/0008-5472.CAN-07-5183.
-
14.
Bergqvist J, Latif A, Roberts SA, Hadfield KD, Lalloo F, Howell A, et al. RASSF1A polymorphism in familial breast cancer. Fam Cancer. 2010;9(3):263-5. [PubMed ID: 20361264]. https://doi.org/10.1007/s10689-010-9335-8.
-
15.
Donninger H, Barnoud T, Nelson N, Kassler S, Clark J, Cummins TD, et al. RASSF1A and the rs2073498 Cancer Associated SNP. Front Oncol. 2011;1:54. [PubMed ID: 22649770]. https://doi.org/10.3389/fonc.2011.00054.
-
16.
Kanzaki H, Hanafusa H, Yamamoto H, Yasuda Y, Imai K, Yano M, et al. Single nucleotide polymorphism at codon 133 of the RASSF1 gene is preferentially associated with human lung adenocarcinoma risk. Cancer Lett. 2006;238(1):128-34. [PubMed ID: 16125301]. https://doi.org/10.1016/j.canlet.2005.07.006.
-
17.
Xiao G, Zhang T, Yao J, Ren J, Cao W, Wu G. The association between RASSF1 gene polymorphisms and lung cancer susceptibility among people in Hubei Province of China. J Huazhong Univ Sci Technolog Med Sci. 2009;29(5):646-9. [PubMed ID: 19821102]. https://doi.org/10.1007/s11596-009-0522-5.
-
18.
Zhou SL, Cui J, Fan ZM, Li XM, Li JL, Liu BC, et al. Polymorphism of A133S and promoter hypermethylation in Ras association domain family 1A gene (RASSF1A) is associated with risk of esophageal and gastric cardia cancers in Chinese population from high incidence area in northern China. BMC Cancer. 2013;13:259. [PubMed ID: 23705663]. https://doi.org/10.1186/1471-2407-13-259.
-
19.
Kawai Y, Sakano S, Okayama N, Suehiro Y, Matsuyama H, Hinoda Y. Association of RASSF1A genotype and haplotype with the progression of clear cell renal cell carcinoma in Japanese patients. BJU Int. 2012;110(7):1070-5. [PubMed ID: 22520501]. https://doi.org/10.1111/j.1464-410X.2012.11143.x.
-
20.
Bayram S. RASSF1A Ala133Ser polymorphism is associated with increased susceptibility to hepatocellular carcinoma in a Turkish population. Gene. 2012;498(2):264-9. [PubMed ID: 22394463]. https://doi.org/10.1016/j.gene.2012.02.040.
-
21.
Meyer A, Coinac I, Bogdanova N, Dubrowinskaja N, Turmanov N, Haubold S, et al. Apoptosis gene polymorphisms and risk of prostate cancer: a hospital-based study of German patients treated with brachytherapy. Urol Oncol. 2013;31(1):74-81. [PubMed ID: 21396839]. https://doi.org/10.1016/j.urolonc.2010.09.011.
-
22.
Shivakumar L, Minna J, Sakamaki T, Pestell R, White MA. The RASSF1A tumor suppressor blocks cell cycle progression and inhibits cyclin D1 accumulation. Mol Cell Biol. 2002;22(12):4309-18. [PubMed ID: 12024041].
-
23.
El-Kalla M, Onyskiw C, Baksh S. Functional importance of RASSF1A microtubule localization and polymorphisms. Oncogene. 2010;29(42):5729-40. [PubMed ID: 20697344]. https://doi.org/10.1038/onc.2010.316.
-
24.
Bayram S. Association between RASSF1A Ala133Ser polymorphism and cancer susceptibility: a meta-analysis involving 8,892 subjects. Asian Pac J Cancer Prev. 2014;15(8):3691-8. [PubMed ID: 24870779].
-
25.
Qu LS, Liu JX, Liu TT, Shen XZ, Chen TY, Ni ZP, et al. Association of hepatitis B virus pre-S deletions with the development of hepatocellular carcinoma in Qidong, China. PLoS One. 2014;9(5). eee98257. [PubMed ID: 24849936]. https://doi.org/10.1371/journal.pone.0098257.
-
26.
Lee JH, Kim HY, Kim YJ, Yoon JH, Chung JW, Lee HS. Barcelona Clinic Liver Cancer staging system and survival of untreated hepatocellular carcinoma in a hepatitis B virus endemic area. J Gastroenterol Hepatol. 2015;30(4):696-705. [PubMed ID: 25250761]. https://doi.org/10.1111/jgh.12788.
-
27.
Burbee DG, Forgacs E, Zochbauer-Muller S, Shivakumar L, Fong K, Gao B, et al. Epigenetic inactivation of RASSF1A in lung and breast cancers and malignant phenotype suppression. J Natl Cancer Inst. 2001;93(9):691-9. [PubMed ID: 11333291].
-
28.
Vera J, Schultz J, Ibrahim S, Raatz Y, Wolkenhauer O, Kunz M. Dynamical effects of epigenetic silencing of 14-3-3sigma expression. Mol Biosyst. 2010;6(1):264-73. [PubMed ID: 20024089]. https://doi.org/10.1039/b907863k.
-
29.
Kastan MB, Lim DS. The many substrates and functions of ATM. Nat Rev Mol Cell Biol. 2000;1(3):179-86. [PubMed ID: 11252893]. https://doi.org/10.1038/35043058.
-
30.
Hamilton G, Yee KS, Scrace S, O'Neill E. ATM regulates a RASSF1A-dependent DNA damage response. Curr Biol. 2009;19(23):2020-5. [PubMed ID: 19962312]. https://doi.org/10.1016/j.cub.2009.10.040.
-
31.
Smith J, Tho LM, Xu N, Gillespie DA. The ATM-Chk2 and ATR-Chk1 pathways in DNA damage signaling and cancer. Adv Cancer Res. 2010;108:73-112. [PubMed ID: 21034966]. https://doi.org/10.1016/B978-0-12-380888-2.00003-0.