Abstract
Background:
Macrophages play critical roles in innate immune response in the liver. Whether macrophages participate in liver innate immunity against HCV replication is poorly understoodObjectives:
The aim of this study was to investigate the role of macrophages in liver innate immunity against HCV replication.Materials and Methods:
Freshly isolated monocytes were purified from peripheral blood of healthy adult donors. Macrophages refer to 7-day-cultured monocytes in vitro. A hepatoma cell line (Huh7) was infected with HCV JFH-1 to generate in vitro HCV infectious system. RT-PCR was used to determine HCV RNA and mRNA levels of genes expression. ELISA was used to measure the protein level of interferon-α (IFN-α) and western blot was used to determine protein expression level of Toll-like receptor 3 (TLR3).Results:
HCV dsRNA induced the expression of type I IFN (IFN-α/β) in monocyte-derived macrophages. HCV dsRNA also induced the expression of TLR3 and IFN regulatory factor-7 (IRF-7), the key regulators of the IFN signaling pathway. When HCV JFH-1-infected Huh7 cells were co-cultured with macrophages activated with HCV dsRNA or incubated in media conditioned with supernatant (SN) from HCV dsRNA-activated macrophages, HCV replication was significantly suppressed. This macrophage SN action on HCV inhibition was mediated through type I IFN, which was evidenced by the observation that antibody to type I IFN receptor could neutralize the macrophages-mediated anti-HCV effect. The role of type I IFN in macrophages-mediated anti-HCV activity is further supported by the observation that HCV dsRNA-activated macrophages SN treatment induced the expression of several IFN-stimulated genes (ISGs), ISG15, ISG56, OAS-1, OAS-2, MxA and Viperin in HCV-infected Huh7 cells.Conclusions:
Macrophages may play an important role in liver innate immunity against HCV replication through a type I IFN-dependent mechanism.Keywords
Hepatitis C Virus Interferon Macrophages Toll-Like Receptor 3
1. Background
Hepatitis C virus (HCV) infection is a major cause of chronic hepatitis, liver cirrhosis and hepatocellular carcinoma (1). The interaction between HCV and host innate immunity plays a key role in the immunopathogenesis of HCV disease. Innate immunity is the first line of defense against viral infections. The host innate immune system recognizes pathogens and responds to their stimuli mainly through pattern recognition receptors (PPRs). Toll-like receptors (TLRs) are key sensors that recognize viral pathogen-associated molecular patterns (PAMPs) during viral infection (2). Several TLR members play a critical role in recognition of viral nucleic acids (3). Among these TLRs, TLR3 has a crucial role in virus-mediated innate immune responses (4, 5), as it recognizes dsRNA (6), which either constitutes the genome of one class of viruses or is generated during the life cycle of many viruses, including HCV (4, 5, 7). Sensing through TLR3 activates IFN signaling pathway and induces the production of type І IFNs (IFN-α/β). IFN- α/β have been recognized as the first line of TLR3 activation-mediated antiviral response (8). Therefore, activation of TLR3 signaling pathway in viral target cells could inhibit virus infections such as herpes simplex virus-1 (HSV-1) (9), HIV (10) and HCV (7).
Although cellular and humoral immune responses are present during acute and chronic HCV infection, they are ineffective in eradicating the virus (11). Most HCV-infected subjects develop chronic infection, suggesting that HCV has evolved the strategies to overcome or evade host immune responses (12). Studies (12) of HCV-host interactions revealed that HCV can use several approaches to compromise the host immune response during viral infection. HCV NS3/4A protease is able to impair TLR3 signaling by cleaving the Toll-IL-1 receptor domain-containing adaptor inducing IFN-β (TRIF) adaptor protein and block RIG-I signaling by cleaving the mitochondrial antiviral signaling protein (MAVS) of the mitochondria to inhibit IRF-3 activation and IFN-β expression (13-15).
Macrophages are phagocytic mononuclear cells of the innate immune response, which also participate in adaptive responses. Resident liver macrophages, Kupffer cells (KCs) constitute 15-20% of the total nonparenchymal cells in the liver (16). KCs sense pathogens and help to maintain the tolerogenic environment of the liver (16). Total macrophage numbers increase significantly in chronic HCV-infected liver, which represents a local proliferation of KCs and infiltration of bone-marrow-derived monocytes from peripheral blood (17). Macrophages are key regulators of early immune responses during viral infection of the liver. Macrophages determine cellular and cytokine milieu in HCV-infected liver and contribute to liver inflammation (18). Furthermore, macrophages influence HCV-induced liver fibrosis, cirrhosis and hepatocellular carcinoma (18). However, the role of macrophages in liver innate immunity against HCV replication is poorly understood.
2. Objectives
In the present study, we aimed to investigate the role of macrophages in liver innate immunity against HCV replication.
3. Materials and Methods
3.1. Reagents
T7 and T3 MegaScript kits were purchased from Ambion, Life Technology (Austin, TX). LyoVec transfection reagent was purchased from Invivogen (San Diego, CA). ELISA kit for human IFN-α was purchased from PBL Assay Science (Piscataway, NJ). Rabbit antibody against TLR3 was purchased from Novus Biologicals (Littleton, CO). Rabbit antibody against GAPDH was purchased from Cell Signaling (Beverly, MA). Mouse anti-IFNAR2 antibody was purchased from LifeSpan BioScience, Inc. (Seattle, WA). Mouse IgG was purchased from Molecular Probes (Eugene, OR).
3.2. Cell Culture
Peripheral blood was purchased from the Center for AIDS Research at the University of Pennsylvania. The protocol used to isolate blood from donors, purify the blood components and distribute this material to the investigators was approved by the IRB of the Center for AIDS Research. These blood samples were screened for all normal blood-borne pathogens and certified to be pathogen free. Monocytes were purified from peripheral blood of three healthy adult donors according to our previously described technique (19). Freshly isolated monocytes were cultured in 48-well culture plates at a density of 2.5 × 105 cells/well in Dulbecco modified Eagle medium (DMEM) containing 10% fetal calf serum. Macrophages refer to 7-day-cultured monocytes in vitro. A hepatoma cell line (Huh7) provided by Dr. Charles Rice (The Rockefeller University, New York, NY, USA) was maintained in DMEM with 10% FBS, penicillin (100 U/mL) and streptomycin (100 μg/mL).
3.3. Plasmids and Virus
The HCV pBSII-core, pBSII-E1-p7, pBSII-HCV-NS-3’NTR and pCMV-NS5A plasmids were generously provided by Dr. Kui Li (University of Tennessee, Memphis, TN, USA) (20). HCV ssRNAs were synthesized from linearized vectors using T7 and T3 MegaScript kits, as described previously (20). To form dsRNA duplexes, equal molar amounts of +ssRNAs and -ssRNAs were combined in a microcentrifuge tube and incubated at 100°C for 5 minutes, followed by a slow cool-down to room temperature (20). The generation of infectious HCV JFH-1 and infection of Huh7 cells (MOI of 0.01) were performed as previously described (21). HCV JFH-1 infection of Huh7 was analyzed by real time RT-PCR for HCV RNA.
3.4. HCV dsRNA Stimulation and co-Culture of Huh7 Cells With Macrophages
Monocyte-derived macrophages (MDM) were stimulated with HCV dsRNA (1 μg/mL) using LyoVec transfection reagent. Cells were collected for total RNA extraction after 24 hours stimulation and supernatant (SN) was collected after 48 hours stimulation. For the co-culture experiments, MDM was first stimulated with HCV dsRNA (1 μg/mL) for 16 hours and then co-cultured with HCV JFH-1-infected Huh7 cells (day 3 postinfection) in 0.4 μm-pore-transwell tissue culture plates (Costar, Cambridge, MA). MDM was placed in the lower compartment and Huh7 cells cultured in the upper compartment. Huh7 cells collected for RNA extraction and real-time RT-PCR 48 hours after co-culture. For the experiments using MDM SN, HCV JFH-1-infected Huh7 cells were cultured in media with or without SN from MDM stimulated with HCV dsRNA (5%, 10% and 20%, vol/vol) for 48 hours. MDM SN was added to Huh7 cells infected with JFH-1 at day 3 postinfection. SN from MDM incubated with LyoVec was only used as a negative control.
3.5. RNA Extraction and Real Time RT-PCR
Total RNA from cultured cells was extracted with Tri-Reagent (Molecular Research Center, Cincinnati, OH) as previously described (22). Total RNA (1 μg) was subjected to RT using the RT system (Promega, Madison, WI) with random primers for one hour at 42°C. The reaction was terminated by incubating the reaction mixture at 99°C for 5 minutes and the mixture was kept at 4°C. The resulting cDNA was used as a template for real time PCR quantification. Real time PCR was performed with 1/10 of the cDNA with the iQ SYBR Green Supermix (Bio-Rad Laboratories, Hercules, CA) as previously described (22). The amplified products were visualized and analyzed using the software MyiQ provided with the thermocycler (iCycler iQ real time PCR detection system; Bio-Rad Laboratories). The oligonucleotide primers were synthesized by Integrated DNA Technologies, Inc. (Coralville, IA), and sequences are shown in Table 1. cDNA was amplified by PCR and the products were measured using SYBR green I (Bio-Rad Laboratories, Inc., Hercules, CA). Data was normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and presented as the change in induction relative to that of untreated control cells.
Primers for Real-Time RT-PCR
| Primer | Sequences (5’ - 3’) |
|---|---|
| GAPDH | |
| Forward | GGTGGTCTCCTCTGACTTCAACA |
| Reverse | GTTGCTGTAGCCAAATTCGTTGT |
| IFN-α | |
| Forward | TTTCTCCTGCCTGAAGGACAG |
| Reverse | GCTCATGATTTCTGCTCTGACA |
| IFN-β | |
| Forward | GCCGCATTGACCATCTATGAGA |
| Reverse | GAGATCTTCAGTTTCGGAGGTAAC |
| TLR3 | |
| Forward | AGCCACCTGAAGTTGACTCAGG |
| Reverse | CAGTCAAATTCGTGCAGAAGGC |
| IRF-7 | |
| Forward | TGGTCCTGGTGAAGCTGGAA |
| Reverse | GATGTCGTCATAGAGGCTGTTGG |
| ISG15 | |
| Forward | GGCTGGGAGCTGACGGTGAAG |
| Reverse | GCTCCGCCCGCCAGGCTCTGT |
| ISG56 | |
| Forward | TTCGGAGAAAGGCATTAGA |
| Reverse | TCCAGGGCTTCATTCATAT |
| OAS-1 | |
| Forward | AGAAGGCAGCTCACGAAACC |
| Reverse | CCACCACCCAAGTTTCCTGTA |
| OAS-2 | |
| Forward | CAGTCCTGGTGAGTTTGCAGT |
| Reverse | ACAGCGAGGGTAAATCCTTGA |
| MxA | |
| Forward | GCCGGCTGTGGATATGCTA |
| Reverse | TTTATCGAAACATCTGTGAAAGCAA |
| Viperin | |
| Forward | TGGGTGCTTACACCTGCTG |
| Reverse | TGAAGTGATAGTTGACGCTGGT |
| HCV | |
| Forward | RAYCACTCCCCTGTGAGGAAC |
| Reverse | TGRTGCACGGTCTACGAGACCTC |
3.6. Enzyme-Linked Immunosorbent Assay
SN collected from HCV dsRNAs-stimulated MDM cell cultures was directly tested for IFN-α protein level by ELISA, which was performed according to the manufacturer’s instructions.
3.7. Western Blot Analysis
The expression of TLR3 and GAPDH protein was evaluated by immunoblot analysis. Following incubation with specific antibodies and extensive washing in PBS containing 0.05% Tween-20, membranes were incubated with horseradish peroxidase-conjugated goat anti-rabbit IgG (Pierce, Chester, UK) for one hour at room temperature. Membranes were extensively washed in PBS containing 0.05% Tween-20, and immunoblots were visualized by Fujifilm Las-1000 Luminescent Image Analyzer (Fujifilm, Tokyo, Japan).
3.8. Statistical Analysis
Student’s t-test was used to evaluate the significance of difference between the groups and multiple comparisons were performed by regression analysis and one-way analysis of variance. P values below 0.05 were considered statistically significant. All data presented as mean ± SD. Statistical analyses were performed using (SPSS Inc., Chicago, IL, USA) for Windows.
4. Results
4.1. HCV dsRNA Induces IFN-α/β Expression
We first examined the effect of HCV dsRNAs on IFN-α/β expression in MDM. MDM was stimulated with 4 HCV dsRNAs (Core, E1-P7, NS-3’NTR and NS5A, 1 μg/mL) for 24 hours, respectively. As shown in Figure 1, Core, E1-P7, NS-3’NTR and NS5A dsRNAs significantly induced IFN-α (Figure 1 A) and IFN-β (Figure 1 B) mRNA expression in MDM. IFN-α protein secretion by HCV dsRNA-stimulated MDM was confirmed by ELISA (Figure 1 C).
Effect of HCV dsRNAs on IFN-α/β Expression in Macrophages

4.2. HCV dsRNA Induces TLR3 and IRF-7 Expression
Investigating the mechanism (s) of HCV dsRNA-mediated induction of IFN-α/β revealed that HCV dsRNAs (Core, E1-P7, NS-3’NTR and NS5A, 1 μg/mL) significantly increased TLR3 (Figure 2 A) and IRF-7 (Figure 2 B) mRNA expression in MDM. Upregulation of TLR3 protein expression in MDM by HCV dsRNA stimulation was also confirmed by Western-blot (Figure 2 C).
Effect of HCV dsRNAs on TLR3 and IRF-7 Expression in Macrophages

4.3. HCV dsRNA-Stimulated MDM Inhibit HCV Replication in Hepatocytes
In HCV inhibition experiments, we first examined whether MDM or SN from MDM cultures (MDM SN) has cytotoxicity effect on Huh7 cells. Little cytotoxic effect was observed in Huh7 cells either co-cultured with MDM stimulated with or without HCV dsRNA or treated with MDM SN (data not shown). We then determined whether MDM stimulated with HCV dsRNA suppresses HCV replication in Huh7 cells. We demonstrated that HCV replication was significantly inhibited in Huh7 cells co-cultured with MDM stimulated with HCV dsRNA (Figure 3 A). In addition, SN from HCV dsRNA-stimulated MDM cultures, when added to HCV JFH-1-infected Huh7 cells, inhibited viral RNA expression in a concentration-dependent manner (Figure 3 B). We further examined anti-HCV activity of MDM SN from HCV dsRNA-stimulated MDM under three different conditions; Huh7 cells were incubated with MDM SN either 24 hours before HCV infection, or simultaneously with HCV infection, or 8 hours after infection. Cells pretreated for 24 hours with 10% (v/v) of MDM SN and then infected had significantly lower levels of HCV RNA than untreated and infected cells (Figure 3 C). Similarly, cells treated with MDM SN and infected simultaneously or after 8 hours HCV JFH-1 infection had significantly lower levels of HCV RNA than the control cells (Figure 3 C).
HCV dsRNA (NS-3’NTR)-Stimulated Macrophages or Supernatant (SN) From Macrophages Culture Suppresses HCV Replication in Huh7 Cells
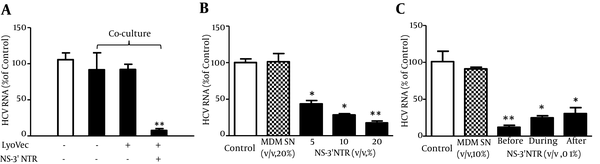
4.4. IFN-α/β are Involved in Anti-HCV Activity of MDM
To investigate whether HCV dsRNA-induced IFN-α/β are responsible for MDM SN-mediated anti-HCV activity, we incubated HCV JFH-1-infected Huh7 cells with antibody to type I IFN receptor (IFNAR2) prior to MDM SN treatment. As shown in Figure 4, antibody to IFNAR2 partially blocked the ability of MDM SN to inhibit HCV replication in Huh7 cells.
Effect of Antibody Against Type I IFN Receptor (IFNAR2) on Macrophages Supernatant (SN)-Mediated anti-HCV Activity
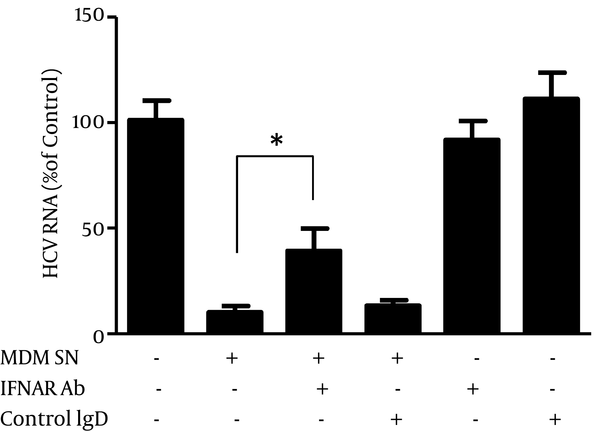
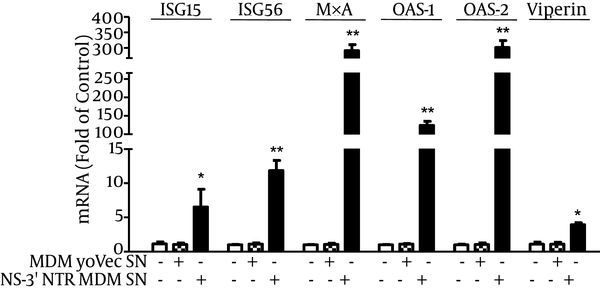
4.5. MDM SN Induces ISGs Expression in HCV-Infected Huh7 Cells
The action of IFN on virus-infected cells is to elicit an antiviral state, which is characterized by induction of IFN-stimulated genes (ISGs) (23). We thus examined the expression of several key ISGs in HCV JFH-1-infected Huh7 cells incubated with SN from MDM cultures stimulated with HCV dsRNA. As shown in Figure 5, SN from HCV dsRNA-stimulated MDM induced ISG15, ISG56, OAS-1, OAS-2, MxA and Viperin gene expression in HCV-infected hepatocytes.
Effect of HCV dsRNA-Activated Macrophages SN on the Expression of ISG15, ISG56, MxA, OAS-1, OAS-2 and Viperin in HCV JFH-1-Infected Huh7 Cells
5. Discussion
In this study, we investigated the role of macrophages in control of HCV replication in hepatocytes. Four HCV dsRNAs (Core, E1-P7, NS-3’NTR and NS5A) were used to investigate the effect of HCV genome on the activation of MDM. Studies (24-26) showed that HCV RNA is highly structured and contains ds regions in various portions of the genome, such as the 5’- and 3’-NTRs and the core- and NS5B-coding regions. Other studies also showed that these four HCV dsRNAs could induce chemokine and inflammatory cytokine expression in hepatocytes (25) and IFNs expression in human type 2 myeloid dendritic cells (26). We showed that HCV dsRNA-activated MDM had the ability to suppress HCV replication in hepatocytes. We observed that HCV dsRNA induced IFN-α/β expression in macrophages. IFN-α/β is the first line of innate immunity against viral infections. We further demonstrated that IFN-α/β were involved in this macrophages-mediated HCV inhibition, which was evidenced by the observation that antibody to type I IFN receptor (IFNAR2) could neutralize the macrophages-mediated anti-HCV effect. In addition, the role of IFN-α/β in macrophages-mediated anti-HCV activity is supported by the observation that macrophages SN treatment induced the expression of ISG15, ISG56, OAS-1, OAS-2, MxA and Viperin in HCV-infected Huh7 cells. These results indicate that macrophages may be a key regulatory bystander, participating in host innate immunity against HCV replication using a type I IFN-dependent mechanism.
Furthermore, HCV dsRNA induced the expression of TLR3 and IRF-7 in macrophages, the key regulators of the IFN signaling pathway. As an important PPR, TLR3 recognizes dsRNA that either constitutes the genome of one class of viruses or is generated during the life cycle of viruses, including HCV dsRNA (20, 27). During the early stages of HCV infection, KCs and infiltrated macrophages are exposed to free viral nucleic acids and proteins. TLR3 expressed in macrophages likely recognizes HCV via phagocytosis and becomes activated, then induces the expression of IFN-α/β and contributes to viral inhibition (18). Here, we used synthesized HCV dsRNA to activated macrophages. We showed for the first time that HCV dsRNA can induce IFN-α/β expression in macrophages in vitro. It is most likely that HCV dsRNA induces IFN-α/β expression by activating TLR3 signaling pathway in macrophages.
Because of a lack of proofreading by the RNA-dependent RNA polymerase (RdRp) and high replication level, HCV presents a high degree of genetic variability. According to this genetic variability, classification of genotypes, subtypes, isolates and quasispecies is allowed (28). HCV variability and of quasispecies dynamics play important roles in infection transmission, mechanisms of chronicity and resistance to antiviral therapy (28). Thus, HCV genetic variabilities may potentially affect the activation of TLR3 signaling pathway in macrophages. One weakness of the current study was that we only used HCV dsRNAs generated from single HCV isolate. Future studies are needed to investigate the effect of HCV dsRNA of different genotypes on the TLR3 activation in macrophages.
As important immune cells, macrophages are key regulators of the early immune responses during viral infection in the liver. Macrophages play very important roles in the immunopathogenesis of HCV-related diseases. It was reported that macrophages participate in determining cellular and cytokine milieu in HCV-infected liver, contribute to liver inflammation and influence HCV-induced liver fibrosis, cirrhosis and hepatocellular carcinoma (18). Recent studies (29) showed that HCV can infect primary human macrophages in vitro, inducing tumor necrosis factor-α (TNF-α) and interleukin 8 (IL-8) expression. Other studies (30) showed that HCV can infect monocytes, macrophages and dendritic cells (DCs) in vivo. It was also showed that high concentrations of HCV particles could stimulate macrophages to express TNF-α and promote HCV entry into polarized hepatoma cells (31). Hepatic macrophages (KCs) link HCV infection with liver inflammation and disease through producing IL-1β (32). It is also showed that serum IL-1β levels are elevated in patients with chronic hepatitis C compared to healthy controls and KCs are the primary cellular source of IL-1β (32). IL-1β drives proinflammatory cytokine, chemokine and immune-regulatory gene expression networks with HCV disease severity (32). KCs can engulf apoptotic bodies of hepatocytes and stimulate death ligand and cytokine expression, which promote liver inflammation and fibrogenesis (33). Furthermore, HCV core and NS3 proteins can activate TLR2 signaling of macrophages and stimulate TNF-α and IL-10 production (34), and TLR1 and 6 are involved in this TLR2-mediated macrophages activation by HCV proteins (35). HCV also can activate inflammasome in monocytes and macrophages through TLR7 signaling pathway in an infection-independent manner (36). In our study, we demonstrated that HCV dsRNA-activated macrophages had the ability to suppress HCV replication in hepatocytes by producing IFN-α/β. Thus, macrophages play roles in both favorable and adverse responses to HCV infection in the liver.
Autophagy plays critical roles in many cellular processes including development, differentiation, survival and homoeostasis. It has been reported that autophagy plays an important role in the HCV-life cycle. HCV induces autophagy and uses autophagy-related proteins (Beclin 1, Atg4B, Atg5 and Atg12) for translation of viral mRNA to initiate its replication (37). Thus, the autophagy pathway can be a target for drugs development for HCV treatment. Although HCV is a hepatotropic virus, there are evidences to support the idea that HCV infection is a multifaceted disease, which causes many extrahepatic manifestations, such as mixed cryoglobulinemia, B-cell-derived non-Hodgkin’s lymphoma (NHL), diabetes mellitus, idiopathic pulmonary fibrosis, autoimmune thyroiditis, sicca syndrome, noncryoglobulinaemic nephropathies, glomerulonephritis and aortic atherosclerosis (38). Macrophage is a target cell of HIV. Because of similar transmission routes, HCV/HIV co-infection is a serious public health problem in high risk groups. It has been reported that antiretroviral therapy (ART) potentially increases the plasma HCV viral load among HCV/HIV co-infected individuals in an ethnic minority area in China (25). Taken together, chronic HCV infection is still a serious global health threat.
In conclusion, our study provided experimental evidences that macrophages may participate in liver innate immunity against HCV replication using a type I IFN-dependent mechanism (Figure 6).
Schematic Diagram of the Mechanisms Involved in Macrophages-Mediated Suppression of HCV Replication in Hepatocytes

Acknowledgements
References
-
1.
Alter MJ. Prevention of spread of hepatitis C. Hepatology. 2002;36(5 Suppl 1):S93-8. [PubMed ID: 12407581]. https://doi.org/10.1053/jhep.2002.36389.
-
2.
Kawai T, Akira S. Toll-like receptor and RIG-I-like receptor signaling. Ann N Y Acad Sci. 2008;1143:1-20. [PubMed ID: 19076341]. https://doi.org/10.1196/annals.1443.020.
-
3.
Kawai T, Akira S. Innate immune recognition of viral infection. Nat Immunol. 2006;7(2):131-7. [PubMed ID: 16424890]. https://doi.org/10.1038/ni1303.
-
4.
Kumar A, Zhang J, Yu FS. Toll-like receptor 3 agonist poly(I:C)-induced antiviral response in human corneal epithelial cells. Immunology. 2006;117(1):11-21. [PubMed ID: 16423036]. https://doi.org/10.1111/j.1365-2567.2005.02258.x.
-
5.
West J, Damania B. Upregulation of the TLR3 pathway by Kaposi's sarcoma-associated herpesvirus during primary infection. J Virol. 2008;82(11):5440-9. [PubMed ID: 18367536]. https://doi.org/10.1128/JVI.02590-07.
-
6.
Alexopoulou L, Holt AC, Medzhitov R, Flavell RA. Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature. 2001;413(6857):732-8. [PubMed ID: 11607032]. https://doi.org/10.1038/35099560.
-
7.
Wang N, Liang Y, Devaraj S, Wang J, Lemon SM, Li K. Toll-like receptor 3 mediates establishment of an antiviral state against hepatitis C virus in hepatoma cells. J Virol. 2009;83(19):9824-34. [PubMed ID: 19625408]. https://doi.org/10.1128/JVI.01125-09.
-
8.
Janeway CAJ, Medzhitov R. Innate immune recognition. Annu Rev Immunol. 2002;20:197-216. [PubMed ID: 11861602]. https://doi.org/10.1146/annurev.immunol.20.083001.084359.
-
9.
Li J, Ye L, Wang X, Hu S, Ho W. Induction of interferon-gamma contributes to Toll-like receptor 3-mediated herpes simplex virus type 1 inhibition in astrocytes. J Neurosci Res. 2012;90(2):399-406. [PubMed ID: 22057682]. https://doi.org/10.1002/jnr.22758.
-
10.
Zhou Y, Wang X, Liu M, Hu Q, Song L, Ye L, et al. A critical function of toll-like receptor-3 in the induction of anti-human immunodeficiency virus activities in macrophages. Immunology. 2010;131(1):40-9. [PubMed ID: 20636339]. https://doi.org/10.1111/j.1365-2567.2010.03270.x.
-
11.
Battaglia AM, Hagmeyer KO. Combination therapy with interferon and ribavirin in the treatment of chronic hepatitis C infection. Ann Pharmacother. 2000;34(4):487-94. [PubMed ID: 10772437].
-
12.
Gale MJ, Foy EM. Evasion of intracellular host defence by hepatitis C virus. Nature. 2005;436(7053):939-45. [PubMed ID: 16107833]. https://doi.org/10.1038/nature04078.
-
13.
Li K, Foy E, Ferreon JC, Nakamura M, Ferreon AC, Ikeda M, et al. Immune evasion by hepatitis C virus NS3/4A protease-mediated cleavage of the Toll-like receptor 3 adaptor protein TRIF. Proc Natl Acad Sci U S A. 2005;102(8):2992-7. [PubMed ID: 15710891]. https://doi.org/10.1073/pnas.0408824102.
-
14.
Foy E, Li K, Sumpter RJ, Loo YM, Johnson CL, Wang C, et al. Control of antiviral defenses through hepatitis C virus disruption of retinoic acid-inducible gene-I signaling. Proc Natl Acad Sci U S A. 2005;102(8):2986-91. [PubMed ID: 15710892]. https://doi.org/10.1073/pnas.0408707102.
-
15.
Li XD, Sun L, Seth RB, Pineda G, Chen ZJ. Hepatitis C virus protease NS3/4A cleaves mitochondrial antiviral signaling protein off the mitochondria to evade innate immunity. Proc Natl Acad Sci U S A. 2005;102(49):17717-22. [PubMed ID: 16301520]. https://doi.org/10.1073/pnas.0508531102.
-
16.
Racanelli V, Rehermann B. The liver as an immunological organ. Hepatology. 2006;43(2 Suppl 1):S54-62. [PubMed ID: 16447271]. https://doi.org/10.1002/hep.21060.
-
17.
Khakoo SI, Soni PN, Savage K, Brown D, Dhillon AP, Poulter LW, et al. Lymphocyte and macrophage phenotypes in chronic hepatitis C infection. Correlation with disease activity. Am J Pathol. 1997;150(3):963-70. [PubMed ID: 9060834].
-
18.
Heydtmann M. Macrophages in hepatitis B and hepatitis C virus infections. J Virol. 2009;83(7):2796-802. [PubMed ID: 18842723]. https://doi.org/10.1128/JVI.00996-08.
-
19.
Hassan NF, Campbell DE, Douglas SD. Purification of human monocytes on gelatin-coated surfaces. J Immunol Methods. 1986;95(2):273-6. [PubMed ID: 3794346].
-
20.
Li K, Li NL, Wei D, Pfeffer SR, Fan M, Pfeffer LM. Activation of chemokine and inflammatory cytokine response in hepatitis C virus-infected hepatocytes depends on Toll-like receptor 3 sensing of hepatitis C virus double-stranded RNA intermediates. Hepatology. 2012;55(3):666-75. [PubMed ID: 22030901]. https://doi.org/10.1002/hep.24763.
-
21.
Zhong J, Gastaminza P, Cheng G, Kapadia S, Kato T, Burton DR, et al. Robust hepatitis C virus infection in vitro. Proc Natl Acad Sci U S A. 2005;102(26):9294-9. [PubMed ID: 15939869]. https://doi.org/10.1073/pnas.0503596102.
-
22.
Yang JH, Lai JP, Douglas SD, Metzger D, Zhu XH, Ho WZ. Real-time RT-PCR for quantitation of hepatitis C virus RNA. J Virol Methods. 2002;102(1-2):119-28. [PubMed ID: 11879700].
-
23.
Katze MG, He Y, Gale MJ. Viruses and interferon: a fight for supremacy. Nat Rev Immunol. 2002;2(9):675-87. [PubMed ID: 12209136]. https://doi.org/10.1038/nri888.
-
24.
Tuplin A, Wood J, Evans DJ, Patel AH, Simmonds P. Thermodynamic and phylogenetic prediction of RNA secondary structures in the coding region of hepatitis C virus. RNA. 2002;8(6):824-41. [PubMed ID: 12088154].
-
25.
Liu X, He N, Fu Z, Duan S, Gao M, Zhang ZF. Plasma Hepatitis C Virus Viral Load Among Hepatitis C Virus Mono-Infected and HCV/HIV Co-Infected Individuals in Yunnan Province,China. Hepat Mon. 2012;12(7):453-9. [PubMed ID: 23008726]. https://doi.org/10.5812/hepatmon.6160.
-
26.
Zhang S, Kodys K, Li K, Szabo G. Human type 2 myeloid dendritic cells produce interferon-lambda and amplify interferon-alpha in response to hepatitis C virus infection. Gastroenterology. 2013;144(2):414-425 e7. [PubMed ID: 23089201]. https://doi.org/10.1053/j.gastro.2012.10.034.
-
27.
Takeda K, Akira S. Toll-like receptors in innate immunity. Int Immunol. 2005;17(1):1-14. [PubMed ID: 15585605]. https://doi.org/10.1093/intimm/dxh186.
-
28.
Argentini C, Genovese D, Dettori S, Rapicetta M. HCV genetic variability: from quasispecies evolution to genotype classification. Future Microbiol. 2009;4(3):359-73. [PubMed ID: 19327119]. https://doi.org/10.2217/fmb.09.8.
-
29.
Radkowski M, Bednarska A, Horban A, Stanczak J, Wilkinson J, Adair DM, et al. Infection of primary human macrophages with hepatitis C virus in vitro: induction of tumour necrosis factor-alpha and interleukin 8. J Gen Virol. 2004;85(Pt 1):47-59. [PubMed ID: 14718619].
-
30.
Revie D, Salahuddin SZ. Role of macrophages and monocytes in hepatitis C virus infections. World J Gastroenterol. 2014;20(11):2777-84. [PubMed ID: 24659871]. https://doi.org/10.3748/wjg.v20.i11.2777.
-
31.
Fletcher NF, Sutaria R, Jo J, Barnes A, Blahova M, Meredith LW, et al. Activated macrophages promote hepatitis C virus entry in a tumor necrosis factor-dependent manner. Hepatology. 2014;59(4):1320-30. [PubMed ID: 24259385]. https://doi.org/10.1002/hep.26911.
-
32.
Negash AA, Ramos HJ, Crochet N, Lau DT, Doehle B, Papic N, et al. IL-1beta production through the NLRP3 inflammasome by hepatic macrophages links hepatitis C virus infection with liver inflammation and disease. PLoS Pathog. 2013;9(4). ee1003330. [PubMed ID: 23633957]. https://doi.org/10.1371/journal.ppat.1003330.
-
33.
Canbay A, Feldstein AE, Higuchi H, Werneburg N, Grambihler A, Bronk SF, et al. Kupffer cell engulfment of apoptotic bodies stimulates death ligand and cytokine expression. Hepatology. 2003;38(5):1188-98. [PubMed ID: 14578857]. https://doi.org/10.1053/jhep.2003.50472.
-
34.
Dolganiuc A, Oak S, Kodys K, Golenbock DT, Finberg RW, Kurt-Jones E, et al. Hepatitis C core and nonstructural 3 proteins trigger toll-like receptor 2-mediated pathways and inflammatory activation. Gastroenterology. 2004;127(5):1513-24. [PubMed ID: 15521019].
-
35.
Chang S, Dolganiuc A, Szabo G. Toll-like receptors 1 and 6 are involved in TLR2-mediated macrophage activation by hepatitis C virus core and NS3 proteins. J Leukoc Biol. 2007;82(3):479-87. [PubMed ID: 17595379]. https://doi.org/10.1189/jlb.0207128.
-
36.
Chattergoon MA, Latanich R, Quinn J, Winter ME, Buckheit R3, Blankson JN, et al. HIV and HCV activate the inflammasome in monocytes and macrophages via endosomal Toll-like receptors without induction of type 1 interferon. PLoS Pathog. 2014;10(5). ee1004082. [PubMed ID: 24788318]. https://doi.org/10.1371/journal.ppat.1004082.
-
37.
Alavian SM, Ande SR, Coombs KM, Yeganeh B, Davoodpour P, Hashemi M, et al. Virus-triggered autophagy in viral hepatitis - possible novel strategies for drug development. J Viral Hepat. 2011;18(12):821-30. [PubMed ID: 22093031]. https://doi.org/10.1111/j.1365-2893.2011.01530.x.
-
38.
Khattab MA, Eslam M, Alavian SM. Hepatitis C virus as a multifaceted disease: a simple and updated approach for extrahepatic manifestations of hepatitis C virus infection. Hepat Mon. 2010;10(4):258-69. [PubMed ID: 22312391].