Abstract
Background:
The ‘a’ determinant domain of hepatitis B surface antigen (HBsAg) (positions 124 to 147) is recognized by antibodies raised either naturally or induced by vaccine. Failure to protect against hepatitis B virus (HBV) infection may occur due to the conformational changes of ‘a’ determinant induced by mutations.Objectives:
The present study analyzed the molecular and three-dimensional (3D) characteristics of the HBsAg ‘a’ determinant mutations among Iranian chronic hepatitis B (CHB) patients, who were vaccine and drug naive.Methods:
Eighty patients with HBsAg positive test results were selected according to the data extracted from questionnaires. Serologic and molecular assays were performed using real-time Polymerase Chain Reaction (PCR) and subsequently surface nested PCR on CHB patients. Next, an extensive mutational analysis was applied following direct sequencing on HBsAg amplified genes. The potential impacts of altered antigenic and 3D properties of amino acid substitutions were carried out using bioinformatics approaches.Results:
All patients were negative for HBeAg and positive for anti-HBe. Mutational analysis showed that 60 (75%) of 80 patients had at least one amino acid substitution. Several mutations were found in ‘a’ determinant (P127L, P127T, G130N, and S136Y). Bioinformatics investigations indicated that all mutations induced a conformational change in ‘a’ determinant region. P127L substitution led to a considerable decreased HBsAg antigenicity compared to other mutants.Conclusions:
The current analyses revealed that the studied mutations induced a local change in the ‘a’ determinant conformation. These findings could be useful for the design of HBsAg detection assays, which may significantly improve the ability to detect particular HBsAg mutants.Keywords
Chronic Hepatitis B Infection Hepatitis B Surface Antigen Mutation Bioinformatics
1. Background
Hepatitis B Virus (HBV) is the agent of a human chronic and carcinogenic infection, which remains a major global health problem. Despite the presence of an effective vaccine since 1982, one-third of the world’s population has been infected with HBV, of whom more than 240 million patients are HBV chronic carriers. These patients are at an extremely high risk for developing end stage liver disease, such as cirrhosis, liver decompensation, Hepatocellular Carcinoma (HCC) (1, 2).
HBsAg composed of 226 amino acids (aa) and contains a highly conformational region from positions 124 to 147, defined as the ‘a’ determinant located in the major hydrophilic region (MHR, aa from 99 to 160) (3-5). Mutations directed to this region lead to conformational changes, rendering the protein being undetectable by some commercially available HBsAg assays (6). Several HBV mutations within the ‘a’ determinant of HBsAg have been reported as being immune escape variants, which could potentially be induced by vaccine or natural immunity in infected individuals (7, 8).
Recognition of this critical region by anti-HBs depends on its three-dimensional (3D) structure, which also relies on the amino acid sequences flanking the ‘a’ determinant (9, 10). To date, the 3D structure of HBsAg has not been fully determined. One of the best ways to alleviate these drawbacks is to employ ‘ab initio’ or ‘de novo’ modeling for the prediction of conformational properties of HBsAg mutants.
2. Objectives
The purpose of the present study was: (i), to explore HBsAg mutations of Iranian HBV treatment-naive chronic carriers (ii), to assess the antigenic properties of HBsAg mutants compared to the wild type, and (iii) to predict the 3D conformations of the HBsAg ‘a’ determinant containing mutants.
3. Methods
3.1. Patients
Eighty HBsAg-positive CHB carrier patients, who were referred to the Iranian hepatitis network (IHC), Tehran, during 2013 to 2014 were included in a cross-sectional study. They had no evidence of co-infection with hepatitis C, hepatitis D, and human immunodeficiency viruses. They were all treatment-naive. None had a history of HBV vaccination, alcohol consumption, autoimmune hepatitis, and drug-induced hepatitis. The diagnosis of chronic HBV was made by clinical, serological, biochemical, radiological, and endoscopic criteria. Informed consent was obtained from all enrolled participants, and a questionnaire was distributed to collect data. The study protocol was approved by the IHC ethical committee.
3.2. Laboratory Tests
Sera were tested using commercially available enzyme-linked immune assay kits, according to the manufacturer’s instructions to test serological markers (HBsAg, HBeAg, and anti-HBe) (Acon, San Diego, CA, USA).
The HBV-DNA was extracted from 200 µL of serum samples using the high pure extraction kit (Roche, Germany) and eluted in 50 to 100 µL, according to the manufacturer’s instructions. Viral load was determined by real-time PCR (Fast-Track Diagnostics, Luxembourg) and was expressed in copies/mL, according to the manufacturer’s instructions. All positive samples were selected for HBsAg gene standard PCR using specific primers, as described previously (11).
3.3. Sequencing and Mutational Analysis
Direct sequencing of HBsAg genes was carried out (Genetic Analyzer ABI- 3130 DNA Sequencer, Foster City, CA, USA) using 2 pmol of appropriate internal primers. The electropherograms were examined visually using a Chromas program. Sequences of the surface genes were aligned using a BioEdit package (version 7.0.9).
The reference sequences for HBV genotypes were obtained from the gene bank database (NCBI). Obtained Sequences were aligned with each other using the BioEdit software. The synonymous (dS) and non-synonymous (dN) substitutions were calculated for all coding sequences.
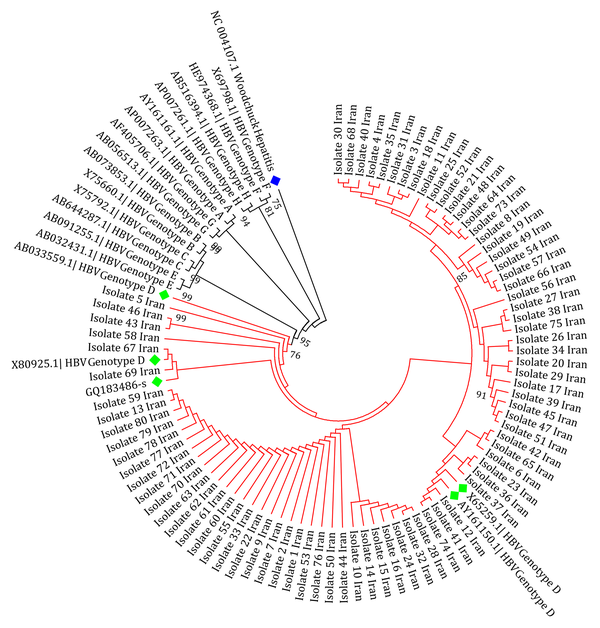
Subtyping was carried out by phylogenetic analysis with reference sequences of HBV genotypes A to H. Phylogenetic tree reconstruction was conducted using molecular evolutionary genetics analysis (MEGA) software (version 6.06), with the maximum likelihood (ML) method in the Kimura two-parameter substitution model with 1000 bootstrap sampling. Significance was based on bootstrap values of 70. The HBsAg gene sequence of the Woodchuck hepatitis virus (WHV) was used as an out-group.
3.4. Evaluation of the Mutations on Conformation of HBsAg
Sequences of the wild-type and mutant types of HBsAg were subjected to the I-TASSER server for ‘ab initio’ prediction of their structures. This server finds appropriate protein fragments of previously solved structures as templates to predict the structures of input protein sequences based on sequence alignment and secondary structure prediction (12). In particular, I-TASSER calculates a normalized B-factor as an extent of thermal mobility of the model by ResQ (an estimated local accuracy of each model), using a combination of template-based assignment and machine-learning-based prediction. Values smaller than zero could be considered as reliable and stable regions (13). Moreover, I-TASSER uses C-score for estimating the quality of predicted models. This score is calculated based on the significance of threading template alignments and the convergence parameters of the structure assembly simulations. In reliable models, the C-score was typically between 2 and 5, where 2 showed the highest confidence.
3.5. Evaluation of Protein Antigenicity
It has been reported that amino acid substitutions both outside and within the ‘a’ determinant of HBsAg could cause a reduced binding affinity to monoclonal antibodies developed against the ‘a’ determinant (14). To assess the antigenicity of the HBsAg mutants, all mutant sequences as well as wild HBsAg were subjected to the antibody epitope prediction server IEDB-AR (15).
3.6. Statistical Analysis
All parameters were evaluated using the statistical package for social Science (SPSS-21, SPSS Inc., Chicago, Illinois, USA). Data were expressed as percentages for categorical variables and means ± SDs for continuous variables.
4. Results
4.1. General Characteristic of the Patients
Eighty CHB patients were enrolled in the study and all were HBV DNA positive and treatment- and vaccine-naive. All patients were negative for HBeAg and were positive for anti-HBe. Demographic and clinical characteristics of the CHB patients are summarized in Table 1. From a total of 80 patients, 60 (75%) and 20 (25%) were male and female, respectively. The ages of patients were between 16 and 70 years old.
Demographic and Clinical Characteristics of the OBI Patients (N = 80)
| Variables | CHB Patients (N = 80) |
|---|---|
| Age, y | 37.29 ± 11.23 |
| Gender, No. (%) | |
| Male | 60 (75) |
| Female | 20 (25) |
| ALT (IU/L) | 121 ± 70.5 |
| AST (IU/L) | 65 ± 51.7 |
| HBV-DNA, Log10 IU/mL | 5.024 ± 1.4 |
| HBeAg, No. (%) | |
| Positive | 0 (0) |
| Negative | 80 (100) |
| Anti-HBe, No. (%) | |
| Positive | 80 (100) |
| Negative | 0 (0) |
4.2. Phylogenetic and Mutational Analysis
Analysis of variation within the HBsAg gene of patients indicated that the only detected genotype and subtype was D (100%) and ayw2 (100%), respectively. The obtained isolates showed a homological pattern in the tree that indicated a slow recent evolution compared to the other reference sequences in the tree (Figure 1). Sixty (75%) of the 80 patients contained at least one amino acid substitution. Overall, out of a total of 240 changes, 102 (42.5%) and 138 (57.5%) were synonymous and non-synonymous, respectively. Twenty (25%) of the HBsAg sequences did not show any nucleotide substitutions. Among 80 samples, 10 (12.5%) contained at least one mutation within the ‘a’ determinant region of HBsAg. The average nucleotide mutation frequency (dS/dN) of all sequences was 0.74, according to the number of mutations per site. A ratio of less than “one” for the HBsAg region indicated a positive selection pressure or in other words, sequence instability.
The Phylogenetic Relationship of Hepatitis B Virus and Other Control Genotypes for the HBsAg Protein
4.3. Mutations in ‘a’ Determinant Region Induce Local Conformational Changes
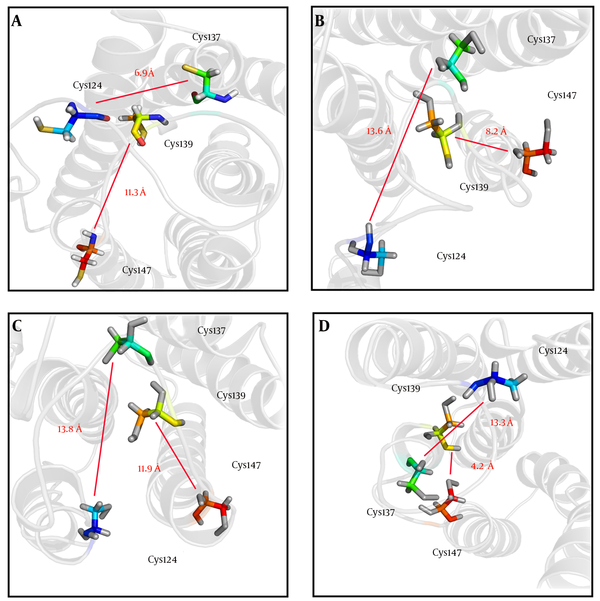
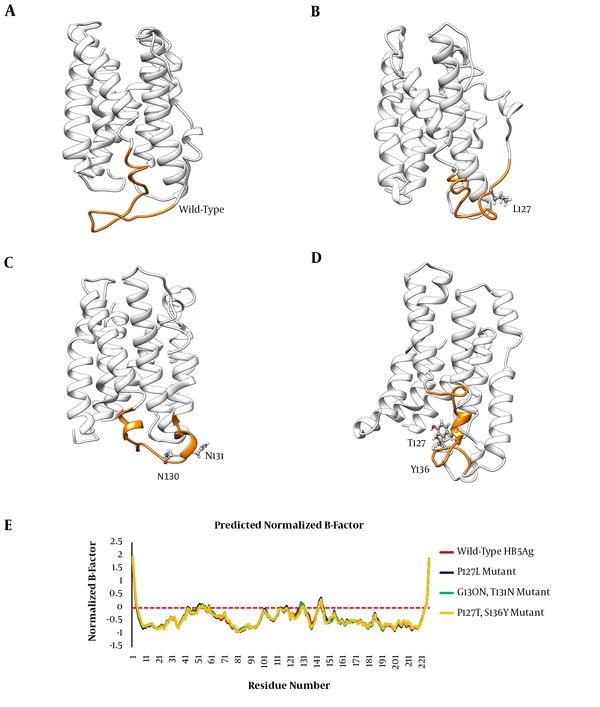
In order to investigate the effects of studied mutations responsible for conformational changes of the HBsAg, the 3D structures of the wild-type and mutant-type proteins were predicted using the I-TASSER server. Results indicated that all mutations induced a local conformational change in the ‘a’ determinant region. However, the effects of HBsAgG130N+T131N and HBsAgP127T+S136Y mutations on the conformation of ‘a’ determinant were more substantial. In addition to induction of local changes in ‘a’ determinant, these mutations could insert an extra α-helix in ‘a’ determinant region of the mutant-type proteins when compared to wild-type HBsAg. The HBsAgG130N, T131N could interfere with the integrity of the HBsAg antigenic loop by probably affecting the distance between cysteine residues 24, 37, 39, and 47, which are necessary for ‘a’ determinant stability. The distance between these cysteine residues was measured by the PyMOL program. As expected, the distance between Cys124 and Cys137 as well as between Cys139 and Cys147 considerably changed upon the studied mutations (Figure 2). Figure 3A - D illustrates the effects of HBsAg mutations on the conformation of ‘a’ determinant region. However, these 3D structures need to be validated to authenticate structures. To this end, B-factor and C-score values were calculated for each predicted model. The C-score of the wild-type, HBsAgP127L, HBsAgG130N+T131N, and HBsAgP127T+S136Y were calculated as -3.24, -4.05, -3.58, and -3.55, respectively, suggesting that the models were acceptable for further analyses. Furthermore, the predicted B-factor values for each HBsAg residue in the modeled structures were plotted. The resulted B-factor plot indicated that the majority of residues had a B-factor value below zero, implicating that the modeled 3D structures of HBsAg were closely related to the native structure. It is a worth noting that residues with B-factor values higher than zero were generally instable in experimentally-characterized structures. However, some large peaks appeared in the ‘a’ determinant region of the protein structures that could be fully attributed to the higher flexibility of this antigenic loop compared to other portions of the protein (Figure 3E).
Calculation of Distances Between Cys124 and Cyc137 as well as Between Cys139 and Cys147 in (A) wild-type HBsAg, (B) HBsAgP127L, (C) HBsAgG130N+T131N and (D) HBsAgP127T+S136Y
Effects of HBsAg Mutations on 3D Structure of the Protein; Predicted 3D Structure of (A) Wild-Type HBsAg, (B) HBsAgP127L, (C) HBsAgG130N+T131N, and (D) HBsAgP127T+S136Y.
4.4. Prediction of Antigenic Index
Mutations in ‘a’ determinant of HBsAg caused a non-significant alteration in the protein antigenicity (Table 2). However, the results of IEDB server revealed that P127L substitution was capable of decreasing HBsAg antigenicity compared with the wild-type sequence (Figure 4B). Also, the HBsAg P127T, S136Y mutant moderately caused a reduction of HBsAg antigenicity in comparison to the wild-type (Figure 4D).
The Effect of Identified Substitutions on HBsAg Antigenicity
| Wild-type | P127L | G130N+T131N | P127T+S136Y | ||||
|---|---|---|---|---|---|---|---|
| Position | Score | Position | Score | Position | Score | Position | Score |
| C124 | 0.902 | C124 | 0.010 | C124 | 0.883 | C124 | 0.622 |
| T125 | 1.098 | T125 | 0.038 | T125 | 1.055 | T125 | 0.423 |
| T126 | 1.206 | T126 | 0.146 | T126 | 1.166 | T126 | 0.832 |
| P127 | 1.301 | L127a | 0.241a | P127 | 1.267 | T127a | 0.926a |
| A128 | 1.461 | A128 | 0.401 | A128 | 1.393 | A128 | 1.086 |
| Q129 | 1.446 | Q129 | 0.385 | Q129 | 1.377 | Q129 | 1.071 |
| G130 | 1.284 | G130 | 0.386 | N130a | 1.216a | G130 | 0.959 |
| T131 | 1.123 | T131 | 0.384 | N131a | 1.054a | T131 | 0.844 |
| S132 | 1.011 | S132 | 0.676 | S132 | 0.943 | S132 | 0.546 |
| M133 | 0.784 | M133 | 0.579 | M133 | 0.698 | M133 | 0.328 |
| Y134 | 0.278 | Y134 | 0.278 | Y134 | 0.253 | Y134 | -0.082 |
| P135 | -0.107 | P135 | -0.107 | P135 | -0.135 | P135 | -0.468 |
| S136 | -0.168 | S136 | -0.168 | S136 | -0.202 | S136a | -0.528a |
The Effect of Identified Mutations on the Antigenicity of HBsAg Variants
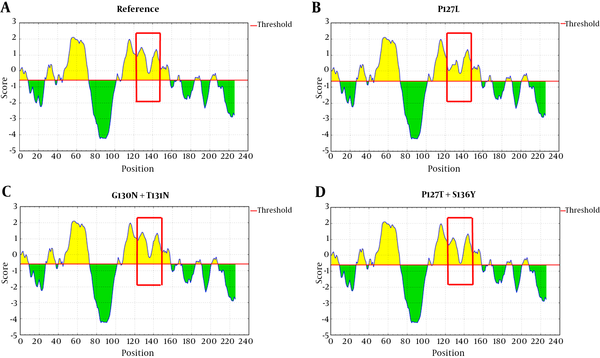
Parameters, such as hydrophilicity, flexibility, accessibility, turns, exposed surface, polarity, and antigenic propensity of polypeptides chains have been correlated with the location of continuous epitopes. obtained obtained from IEDB provide values of calculated scores for each residue. A larger score for the residues might be interpreted as the residue having a higher probability to be a part of the epitope.
5. Discussion
A wide range of varaitions containing various amino acid substitutions across the ‘a’ determinant has been described in the literature (7, 14, 16-18). Amino acid substitutions within the critical positions of structural proteins may influence their stability and structure and also effect their characteristics and interactions with other proteins. Several studies have revealed that the conformation of proteins plays a critical role in their biological activity and interactions with other molecules (19). Previous investigations have indicated that the antigenic determinants of the HBsAg are highly conformation-dependent; therefore, any alteration in the ‘a’ determinant region could interrupt the antigenic index of this protein (20). In the current study, the researchers aimed at investigating the conformational analysis of the HBsAg in the ‘a’ determinant region of Iranian CHB patients, who had not received either active/passive immunization or antivirals.
All patients were infected with genotype D and subtype ayw2. The ratio between silent (synonymous) and missense (non-synonymous) nucleotide mutations (dS/dN) in samples was 0.74, indicating that the proportion of deduced amino acid changes in CHB patients followed a positive selection pattern. The researchers claimed that the emergence of HBsAg mutants, especially in MHR may be associated with modifications on the selective pressures due to the impairment of the immune system in these patients. In fact, the host immune response alone was able to stimulate the selection of HBV variants (21, 22).
It has already been shown that mutations of ‘a’ determinant loops could effect the proper folding of HBsAg (5). The majority of these mutation patterns indicated that the ‘a’ determinant was remarkably affected by these variations. Figure 3 indicated that the structure of HBsAg ‘a’ determinant in the presence of P127L mutation was somehow similar to the reference structure. It is worth noting that both Proline (P) and Leucine (L) are nonpolar amino acids, both of which tend to preserve the space between adjacent residues. In the current study, the researchers found a nonpolar-to-polar (P127T and G130N) mutation that seemed to be involved in a significant change of structure and function of HBsAg. Our computational analysis revealed that HBsAgG130N+T131N and HBsAgP127T+S136Y substitution led to an insertion of, so-called an “extra” α-helix in the ‘a’ determinant region. Glycine (G) is an amino acid with a non-polar side chain, while Asparagine (N) has a more polar and significantly bulkier side chain. Thus, differences in the physicochemical properties of these residues may effect the tree-dimensional conformation and therefore may alter the steric hindrance between other residues (23). The G145R mutation was not observed in the current study. However, previous analyses of the G145R mutant indicated that this mutation inserted a new β-strand at the 121 to 124 position (24). The hydrophilic characteristic of the ‘a’ determinant provides flexibility of its antigenic loops for its interaction with antibodies. Therefore, mutations that impair this 3D flexible structure could potentially impair the interaction between HBsAg and anti-HBs. Although mutations in ‘a’ determinant of HBsAg caused a non-significant alteration in protein antigenicity, the effect of P127L mutation was meaningful (Figure 4).
One of the major limitations of the present study was the lack of in vitro investigation of the phenotypic pattern of each variant. The proposed model for the modified HBsAg structure was based on the software and no experimental study has been carried out to determine the exact modification of structures.
Taken together, a high prevalence of naturally occurring HBsAg mutants (P127L, P127T, G130N, and S136Y) were observed in Iranian CHB treatment and vaccine naive patients. Protein modelling of HBsAg mutants might give insight into the structural basis of these HBV variations at the molecular levels. These findings could be useful for a better understanding of HBV antigen/antibody interactions and for designing novel HBsAg detection assays, which may substantially improve the accuracy of detection of particular HBsAg mutants.
Acknowledgements
References
-
1.
Lavanchy D. Hepatitis B virus epidemiology, disease burden, treatment, and current and emerging prevention and control measures. J Viral Hepat. 2004;11(2):97-107. [PubMed ID: 14996343]. https://doi.org/10.1046/j.1365-2893.2003.00487.x.
-
2.
European Association for the Study of the Liver. Electronic address EEE, European Association for the Study of the L. EASL 2017 Clinical Practice Guidelines on the management of hepatitis B virus infection. J Hepatol. 2017;67(2):370-98. [PubMed ID: 28427875]. https://doi.org/10.1016/j.jhep.2017.03.021.
-
3.
Stirk HJ, Thornton JM, Howard CR. A topological model for hepatitis B surface antigen. Intervirology. 1992;33(3):148-58. [PubMed ID: 1500275]. https://doi.org/10.1159/000150244.
-
4.
Tiollais P, Pourcel C, Dejean A. The hepatitis B virus. Nature. 1985;317(6037):489-95. [PubMed ID: 2995835]. https://doi.org/10.1038/317489a0.
-
5.
Purdy MA. Hepatitis B virus S gene escape mutants. Asian J Transfus Sci. 2007;1(2):62-70. [PubMed ID: 21938236]. https://doi.org/10.4103/0973-6247.33445.
-
6.
Raimondo G, Allain JP, Brunetto MR, Buendia MA, Chen DS, Colombo M, et al. Statements from the Taormina expert meeting on occult hepatitis B virus infection. J Hepatol. 2008;49(4):652-7. [PubMed ID: 18715666]. https://doi.org/10.1016/j.jhep.2008.07.014.
-
7.
Alavian SM, Carman WF, Jazayeri SM. HBsAg variants: diagnostic-escape and diagnostic dilemma. J Clin Virol. 2013;57(3):201-8. [PubMed ID: 22789139]. https://doi.org/10.1016/j.jcv.2012.04.027.
-
8.
Norouzi M, Ghorashi S, Abedi F, Nejatizadeh A, Ataei B, Malekzadeh R, et al. Identification of Hepatitis B Virus Surface Antigen (HBsAg) Genotypes and Variations in Chronic Carriers from Isfahan Province, Iran. Iran J Public Health. 2012;41(3):104-11. [PubMed ID: 23113154].
-
9.
Ie SI, Thedja MD, Roni M, Muljono DH. Prediction of conformational changes by single mutation in the hepatitis B virus surface antigen (HBsAg) identified in HBsAg-negative blood donors. Virol J. 2010;7:326. [PubMed ID: 21087462]. https://doi.org/10.1186/1743-422X-7-326.
-
10.
Carman WF, Zanetti AR, Karayiannis P, Waters J, Manzillo G, Tanzi E, et al. Vaccine-induced escape mutant of hepatitis B virus. Lancet. 1990;336(8711):325-9. [PubMed ID: 1697396]. https://doi.org/10.1016/0140-6736(90)91874-A.
-
11.
Khedive A, Norouzi M, Ramezani F, Karimzadeh H, Alavian SM, Malekzadeh R, et al. Hepatitis B virus surface protein mutations clustered mainly in CTL immune epitopes in chronic carriers: results of an Iranian nationwide study. J Viral Hepat. 2013;20(7):494-501. [PubMed ID: 23730843]. https://doi.org/10.1111/jvh.12045.
-
12.
Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics. 2008;9:40. [PubMed ID: 18215316]. https://doi.org/10.1186/1471-2105-9-40.
-
13.
Yang J, Zhang Y. Protein Structure and Function Prediction Using I-TASSER. Curr Protoc Bioinformatics. 2015;52:5 8 1-15. [PubMed ID: 26678386]. https://doi.org/10.1002/0471250953.bi0508s52.
-
14.
Dindoost P, Jazayeri SM, Karimzadeh H, Saberfar E, Miri SM, Alavian SM. HBsAg variants: Common escape issues. Jundishapur J Microbiol. 2012;5(4):521-7. https://doi.org/10.5812/jjm.4243.
-
15.
Zhang Q, Wang P, Kim Y, Haste-Andersen P, Beaver J, Bourne PE, et al. Immune epitope database analysis resource (IEDB-AR). Nucleic Acids Res. 2008;36(Web Server issue):W513-8. [PubMed ID: 18515843]. https://doi.org/10.1093/nar/gkn254.
-
16.
Carman WF. The clinical significance of surface antigen variants of hepatitis B virus. J Viral Hepat. 1997;4 Suppl 1:11-20. [PubMed ID: 9097273]. https://doi.org/10.1111/j.1365-2893.1997.tb00155.x.
-
17.
Waters JA, Kennedy M, Voet P, Hauser P, Petre J, Carman W, et al. Loss of the common "A" determinant of hepatitis B surface antigen by a vaccine-induced escape mutant. J Clin Invest. 1992;90(6):2543-7. [PubMed ID: 1281839]. https://doi.org/10.1172/JCI116148.
-
18.
Wu C, Deng W, Deng L, Cao L, Qin B, Li S, et al. Amino acid substitutions at positions 122 and 145 of hepatitis B virus surface antigen (HBsAg) determine the antigenicity and immunogenicity of HBsAg and influence in vivo HBsAg clearance. J Virol. 2012;86(8):4658-69. [PubMed ID: 22301154]. https://doi.org/10.1128/JVI.06353-11.
-
19.
Ponsel D, Bruss V. Mapping of amino acid side chains on the surface of hepatitis B virus capsids required for envelopment and virion formation. J Virol. 2003;77(1):416-22. [PubMed ID: 12477846]. https://doi.org/10.1128/JVI.77.1.416-422.2003.
-
20.
Gomez-Gutierrez J, Rodriguez-Crespo I, Gonzalez-Ros JM, Ferragut JA, Paul DA, Peterson DL, et al. Thermal stability of hepatitis B surface antigen S proteins. Biochim Biophys Acta. 1992;1119(3):225-31. [PubMed ID: 1372181]. https://doi.org/10.1016/0167-4838(92)90206-S.
-
21.
Carman WF, Korula J, Wallace L, MacPhee R, Mimms L, Decker R. Fulminant reactivation of hepatitis B due to envelope protein mutant that escaped detection by monoclonal HBsAg ELISA. Lancet. 1995;345(8962):1406-7. [PubMed ID: 7539089]. https://doi.org/10.1016/S0140-6736(95)92599-6.
-
22.
Yamamoto K, Horikita M, Tsuda F, Itoh K, Akahane Y, Yotsumoto S, et al. Naturally occurring escape mutants of hepatitis B virus with various mutations in the S gene in carriers seropositive for antibody to hepatitis B surface antigen. J Virol. 1994;68(4):2671-6. [PubMed ID: 8139044].
-
23.
Lehninger AL, Nelson DL, Cox MM. Amino Acids and Peptides. Lehninger principles of biochemistry. New York: Worth Publishers; 1993. p. 111-33.
-
24.
Rezaee R, Poorebrahim M, Najafi S, Sadeghi S, Pourdast A, Alavian SM, et al. Impacts of the G145R Mutation on the Structure and Immunogenic Activity of the Hepatitis B Surface Antigen: A Computational Analysis. Hepat Mon. 2016;16(7). e39097. [PubMed ID: 27642350]. https://doi.org/10.5812/hepatmon.39097.
