1. Background
Approximately, 257 million people are suffering from chronic infections with hepatitis B virus (HBV) worldwide, and about 887000 of these infected individuals die annually from end-stage hepatic diseases (1, 2). Chronic HBV infection also increases the risk of hepatocellular carcinoma (HCC), which is the fifth most prevalent malignancy and the third cause of cancer-related mortality (3, 4). The incidence of the disease is more than 700,000 cases annually (5), and approximately 50% of the HCC patients suffer from HBV infection (6). Carcinogens, such as aflatoxin or some other chemicals, are believed to act as cofactors for the development of this cancer (7). The HBV x protein (HBx) has a key role in the carcinogenesis associated with the virus, and transgenic mouse models harboring the HBx gene may develop liver cancer (8). HBx induces its effects by transcriptional activation, modulation of apoptosis, deregulation of cell cycle progression, and inhibition of repair of damaged cellular DNA (9). This protein is capable of activating the Notch1 signaling pathway, which may further lead to the development and progression of HCC. Therefore, treatments that decrease Notch1 expression may have anti-tumor effects (10-15).
One characteristic feature of HCC is that effective therapeutic strategies, including surgery and liver transplantation, are only effective in the first stages of tumor formation (16). HCC is a clinically and pathologically heterogeneous disease, so monitoring of patients for early detection of the malignancy is of great importance (16). Although many diagnostic methods including transabdominal ultrasound and surveillance by contrast-enhanced computed tomography are available for the diagnosis of HCC, these methods are not cost-effective and their validity is dependent on the quality of the machines and the experience of the examiner. On the other hand, conventional biomarkers such as α-fetoprotein (AFP), Des-γ-carboxyprothrombin (DCP), and Glypican-3 (GPC3) do not have adequate sensitivity and specificity for early diagnosis (17). Therefore, the use of specific biomarkers that are sensitive enough for the early detection of HCC is crucial for prompt diagnosis (6). Several studies have reported the involvement of microRNAs (miRNAs) in the carcinogenesis and prognosis of HCC (18, 19). MiRNAs are small (18 - 24 nucleotide long) non-coding molecules that are post-transcriptional gene expression regulators and have important roles in many essential biological processes (20). In different types of cancer, the expression of miRNAs changes, and they may act as either oncogenes or tumor suppressors (21). Several studies have revealed a range of miRNAs that regulate liver functions (22). These molecules are stable in biological fluids, such as serum, plasma, urine, semen, amniotic fluid, and CSF (21), and are highly resistant to RNases in body fluids. Currently, they have found the potential to serve as biomarkers for the diagnosis and prognosis of HCC (23).
2. Objectives
In the present study, bioinformatics tools were applied to predict the most important hepatic miRNAs that interact with Notch1 and HBx gene products in humans and HBV, respectively. The plasma levels of the predicted miRNAs were compared in individuals with HBV-induced HCC and control groups using RT-qPCR. These miRNAs were also evaluated as potential biomarkers for the diagnosis of HCC.
3. Methods
3.1. miRNA Prediction Using Bioinformatics Tools
The nucleotide sequences of Notch1 and HBx genes were acquired from NCBI (http://www.ncbi.nlm.nih.gov/gene). The miRNAs that target Notch1 were predicted using different bioinformatics algorithms and tools, including Targetscan, DIANA, miRWalk, miRBase, PicTar, Miranda, microcosm target, and miRNApath. The HBx-targeting miRNAs were predicted using databases such as miRBase and Vir-Mir. After selecting miRNAs that targeted Notch1 and HBx with the highest probability, three miRNAs of the nine predictors for HBx and one miRNA of the five predictors for Notch1 were chosen for further experimental analyses.
3.2. Primer Design
The sequences of the predicted miRNAs were obtained from miRBase. The expression of miR-103 served as the reference target for the normalization of miRNAs expressions. Reverse transcription-specific stem-loop primers and miRNA specific primers (24) were designed using the AlleleID7 and GeneRunner software. The Mfold database was also used to assess the secondary structures. NCBI BLASTn tool was used to evaluate the specificity of the designed primers. The sequences of the primers are presented in Table 1.
| Target | Primer Sequences (5’ - 3’) |
|---|---|
| miR-214 | Sa: 5’-GTCGTATCGAGAGCAGGGTCCGAGGTATTCGCACTCGATACGACACTGCC-3’ |
| Fb: 5’-ACAGCAGGCACAGACAGGCAGT-3’ | |
| miR-6510 | S: 5’- GTCGTATCGAGAGCAGGGTCCGAGGTATTCGCACTCGATACGACCTGCAG-3’ |
| F: 5’-CACCGACTCTGTCTCCTGCAG-3’ | |
| miR-5193 | S: 5’-GTCGTATCGAGAGCAGGGTCCGAGGTATTCGCACTCGATACGACACTGGG-3’ |
| F: 5’-TCCTCCTCTACCTCATCCCAGTG-3’ | |
| miR-34a | S: 5’-GTCGTATCGAGAGCAGGGTCCGAGGTATTCGCACTCGATACGACACAACCA-3’ |
| F: 5’-TGGCAGTGTCTTAGCTGGTTG-3’ | |
| miR-103 | S: 5’-GTCGTATCGAGAGCAGGGTCCGAGGTATTCGCACTCGATACGACCAAGGCA-3’ |
| F: 5’-GCTTCTTTACAGTGCTGCC-3’ | |
| Common reverse | 5’-AGAGCAGGGTCCGAGGT-3’ |
aStem-loop miRNA-specific reverse-transcriptase primers.
bSpecific forward primers for RT-qPCR amplification.
3.3. Patients and Samples
A total number of 73 plasma samples, including 23 samples from individuals with HBV-related HCC, 25 from healthy individuals, and 25 from patients with chronic HBV who had no sign of HCC, were used in this survey. The HCC specimens were collected from the Liver and Digestion Research Center (Taleghani Hospital, Tehran, Iran). These samples had been taken from 10 females and 13 males, with a mean age of 57 years (SD: 8.9; range: 44 to 72 years), not receiving cancer-related treatment including chemotherapy, radiotherapy, or surgery. The chronic HBV samples were collected from the Imam Reza Clinic (Arak University of Medical Sciences, Arak, Iran) and had been obtained from patients suffering from chronic HBV for more than 15 years with no sign of HCC. These samples had been collected from 12 females and 13 males, with a mean age of 48 years (SD: 10.2; range: 39 to 75 years). The samples from healthy individuals, including 12 females and 13 males, with a mean age of 49 years (SD: 10.1; range: 40 to 74 years), were obtained from the Arak Reference Laboratory (Arak University of Medical Sciences, Arak, Iran). All samples were stored at -70°C prior to analysis. The study was approved by the Ethics Committee of Arak University of Medical Sciences (ethics number: 1394.207).
3.4. miRNA Extraction and Reverse Transcription
MicroRNAs were purified from plasma using RNX-Plus kit (SinaClon, Iran) with some modifications to the manufacturer’s protocol. For cDNA synthesis, about 1 µg of RNA was mixed with 1 μM of specific stem-loop RT primers and incubated at 75°C for 5 minutes. A mix containing 100 units of M-MuLV RT-enzyme (Vivantis, Malaysia), 1x RT-buffer, and 400 µM of dNTP were added to the tube. Finally, the RT reaction mixtures were incubated at 25°C for 15 minutes, 37°C for 15 minutes, 42°C for 45 minutes, and 75°C for 10 minutes in a thermal cycler (Eppendorf, Germany). The resulting cDNAs were stored at -20°C prior to RT-qPCR analyses.
3.5. Quantitative Real-time PCR
All RT-qPCR analyses were performed using a LightCycler 96 instrument (Roche, Germany) in triplicates. The reaction mixture contained 7.5 µL of 1x SYBR Green PremixExRaq II (Yekta Tajhiz Azma, Iran), 0.45 µL of sense primer (10 µM concentration), 0.45 µL of antisense primer (10 µM concentration), 1.5 µL of cDNA template, and 5.1 µL of nuclease-free water. The thermal cycling condition was as follows: 95°C for 3 minutes, 40 cycles of 95°C for 10 seconds, 54°C for 10 seconds, and 72°C for 15 seconds. Melting curve analysis was carried out following amplification with a ramp rate of 0.2°C/sec to 96°C with constant fluorescence detection.
3.6. Statistical Analyses
The relative expression was analyzed by the comparative Cq method using the relative expression software tool (REST) (25). RT-qPCR data were expressed as means ± SE. Receiver operating characteristic (ROC) curve analysis was used to estimate the diagnostic capability of miRNA levels. The ROC analysis was performed using SPSS software (version 16). P values of < 0.05 were defined as statistical significance.
4. Results
4.1. miRNA Prediction Results
Based on the predicted results of the Miranda, miRBase, PicTar, TargetScan, miRWalk, and DIANA algorithms, the miRNAs with the highest scores were defined as Notch1-targeting miRNAs (Table 2). Among these miRNAs, miR-34a, which was a consensus target in most of the databases, was selected as the targeting miRNA. The prediction results for HBx-targeting miRNAs using miRBase and MirVir (Table 3) showed that miR-5193, miR-214, and miR-6510 had the highest scores and lowest E-values. In addition, they had the best complementarities with the target gene, so they were selected for further evaluation.
| Gene Name | miRNA | miRWalk | PicTar | Miranda | Targetscan | miRBase | DIANA | SUM |
|---|---|---|---|---|---|---|---|---|
| NOTCH1 | has-miR-34a | 1 | 1 | 1 | 1 | 0 | 1 | 5 |
| NOTCH1 | has-miR-200b | 0 | 0 | 1 | 1 | 0 | 1 | 3 |
| NOTCH1 | has-miR-34c | 0 | 1 | 1 | 1 | 0 | 0 | 3 |
| NOTCH1 | has-miR-139 | 1 | 1 | 1 | 1 | 0 | 0 | 4 |
| NOTCH1 | has-miR-449a | 1 | 0 | 1 | 1 | 0 | 1 | 4 |
| NOTCH1 | has-miR-200c | 0 | 0 | 1 | 0 | 0 | 0 | 1 |
| Gene Name | Predicted miRNA by miRBase | E Value | Score | Predicted miRNA by miRVir | E Value | Score |
|---|---|---|---|---|---|---|
| HBx | hsa-miR-5193 | 1.6 | 74 | miR-3180 | 2.8 | 66 |
| HBx | hsa-miR-214 | 1.9 | 73 | miR-7114 | 6.0 | 62 |
| HBx | hsa-miR-6510 | 6 | 67 | miR-6510 | 7.2 | 61 |
| HBx | hsa-let-7a-2 | 8.8 | 65 | miR-6744 | 7.2 | 61 |
4.2. Quantification of miRNA Plasma Levels
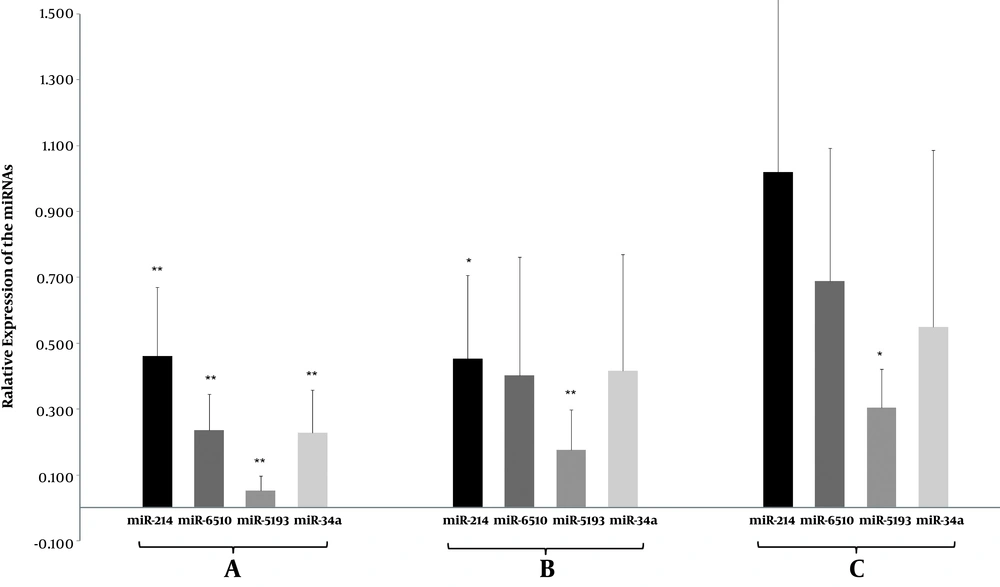
As summarized in Figure 1, the RT-qPCR analysis showed that the expression level of miR-214 was significantly lower in the HCC group than in healthy controls (0.461, P = 0.008) and patients with chronic HBV (0.452, P = 0.023). Similarly, the expression levels were 0.236 (P = 0.001) and 0.402 (P = 0.47) for miR-6510, 0.053 (P = 0.000) and 0.175 (P = 0.000) for miR-5193, and 0.228 (P = 0.003) and 0.416 (P = 0.143) for miR-34a, respectively (Figure 1).
Differential expression levels of miR-214, miR-650, miR-5193, and miR-64a. A, comparison between HCC patients and healthy individuals; B, comparison between HCC patients and chronic HBV patients; C, comparison between chronic HBV patients and healthy individuals. Error bars indicate the standard error of the mean. *P < 0.05, **P < 0.01, significantly different from the control group.
The expression levels of the four miRNAs were also compared between healthy individuals and patients with chronic HBV. The plasma level of miR-5193 (0.304, P = 0.012) was significantly lower in patients with chronic HBV, whereas the levels of miR-214 (1.02, P = 0.96), miR-6510 (0.688, P = 0.208), and miR-34a (0.549, P = 0.179) exhibited no statistical differences between the two groups (Figure 1).
4.3. Determination of Expression Level Cutoff
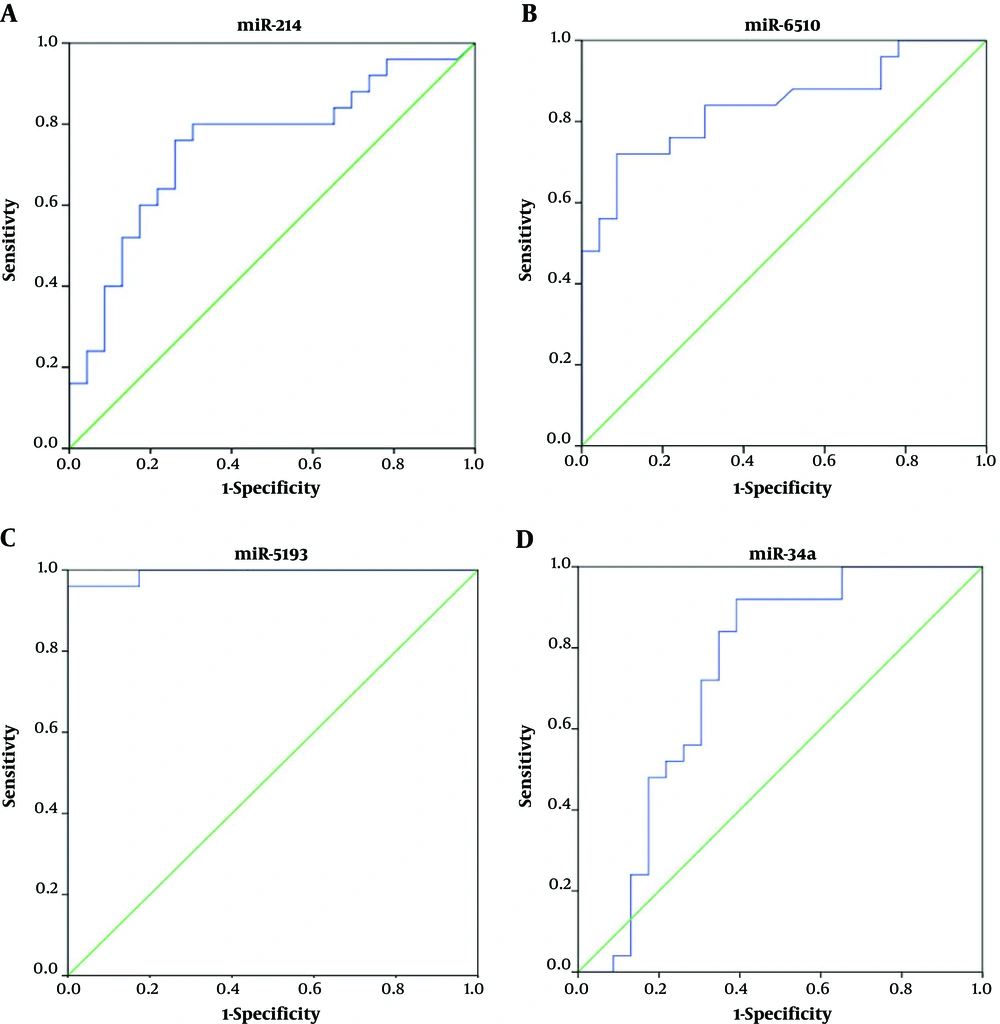
ROC curve analyses were performed to determine the cutoff values of miR-214, miR-6510, miR-5193, and miR-34a, to differentiate HCC patients from healthy individuals and patients with chronic HBV infection. The cutoff levels on the ROC curves were selected using the Youden’s index ([sensitivity + specificity] -1). Comparisons of the miRNA plasma levels between HCC patients and healthy individuals showed that at the cutoff level of 0.792, miR-214 had 76% sensitivity and 74% specificity with an area under the curve (AUC) of 0.747. For miR-6510, at the cutoff level of 0.839, these values were 72% sensitivity and 91% specificity, with an AUC of 0.839. The best sensitivity and specificity were observed with miR-5193, which, at the cutoff point of 0.266, had 96% sensitivity, 100% specificity, and an AUC of 0.993. At the cutoff point of 0.446, miR-34a exhibited 92% sensitivity and 60% specificity, with an AUC of 0.736 (Figure 2).
ROC curve analyses corresponding to the plasma expression of the four miRNAs to discriminate patients with HCC from healthy individuals. A, the area under the curve of miR-214; B, the area under the curve of miR-6510; C, the area under the curve of miR-5193; D, the area under the curve of miR-34a.
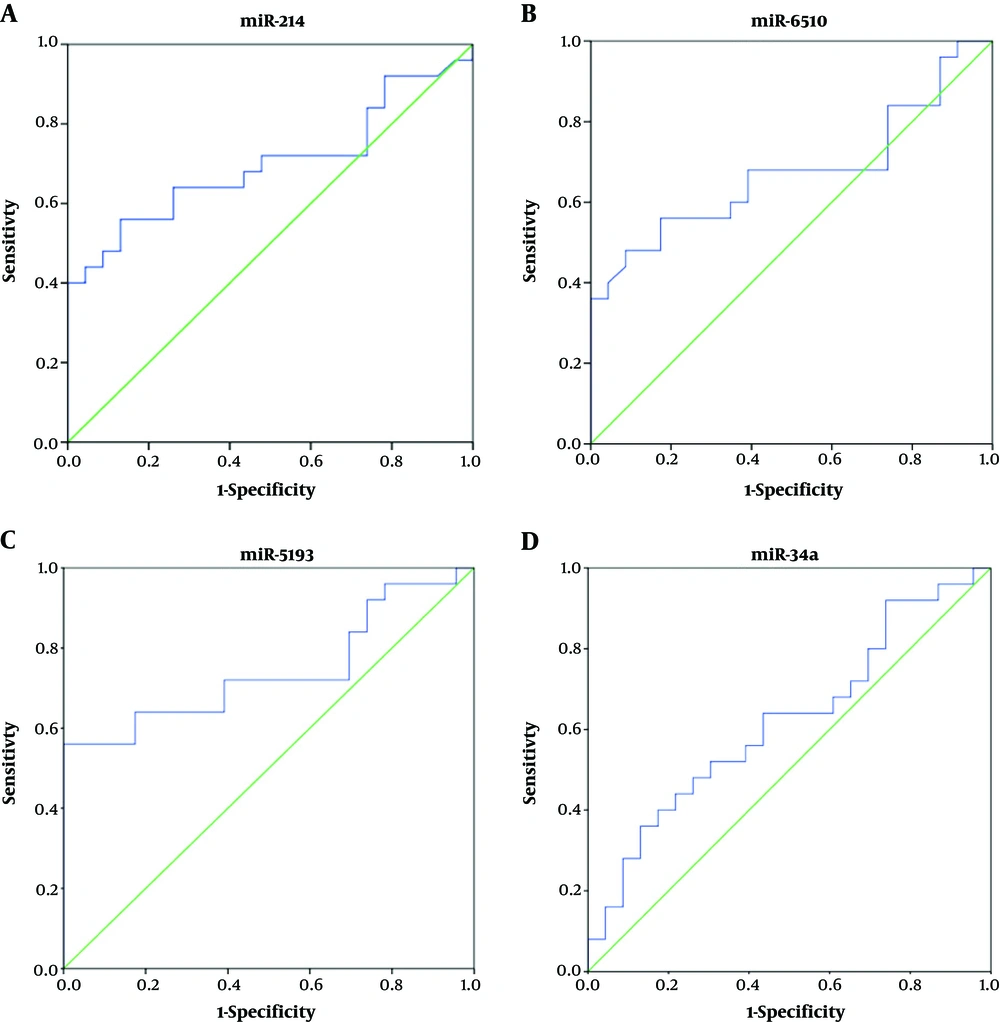
On the other hand, in the comparison between HCC patients and chronic HBV patients, at the cutoff point of 0.806, the sensitivity, specificity, and AUC for miR-214 were 85%, 43%, and 0.52, respectively. The cutoff point of 0.914 for miR-6510 had a sensitivity of 81%, specificity of 39%, and AUC of 0.531. For miR-5193, at the cutoff point of 0.612, these values were 79.8% sensitivity and 82% specificity, with an AUC of 0.817. For miR-34a, the cutoff level of 0.584 exhibited 40% sensitivity and 87% specificity, with an AUC of 0.619 (Figure 3).
ROC curve analyses corresponding to the plasma expression of the four miRNAs to discriminate patients with HCC from patients with chronic HBV. A, the area under the curve of miR-214; B; the area under the curve of miR-6510; C, the area under the curve of miR-5193; D, the area under the curve of miR-34a.
5. Discussion
Several studies have shown that alterations in microRNAs expression are related to the development and progression of various types of human malignancies. The profiles of these miRNAs vary depending on the tumor type, making them novel biomarkers for the diagnosis of different types of cancer including HCC (26). Chronic infection with HBV is one of the main risk factors for the onset and development of HCC, and the viral HBx protein could contribute to a multistep transformation process by affecting specific signal transduction pathways. A few studies have indicated that HBx can dysregulate important signaling pathways, including Notch1, and that Notch1 signaling dysregulation is strongly related to HCC progression (10, 12, 14).
Most studies concerning the detection of miRNAs in cancers have been conducted on tissue samples as an invasive method for cancer diagnosis (27). Several reports have shown the stability of miRNAs in plasma and serum, which might be due to their protection by high-density lipoproteins, exosomes, microvesicles, and Argonaute 2 (28). Therefore, circulating miRNAs could be useful as novel biomarkers for cancer diagnosis or staging. The focus of the present study was, therefore, on the comparative expression of miR-214, miR-6510, miR-5193, and miR-34a using RT-qPCR in the sera of patients with HCC and patients with chronic hepatitis B. These miRNAs were chosen using several algorithms used for the prediction of mRNA-miRNA interactions.
The plasma levels of these four miRNAs, as determined by RT-qPCR, were significantly lower in the HCC group than in the healthy control group. The expression of miR-214, miR-6510, miR-5193, and miR-34a was downregulated by -2.17, -4.23, -18.86, and -4.38 folds, respectively. Comparisons with chronic hepatitis B patients revealed no significant difference in the expression levels of miR-34a and miR-6510. The expression level of miR-5193, on the other hand, was downregulated by -5.71 folds in the HCC group compared to chronic HBV patients. The comparison of the expression levels between the healthy control and chronic HBV groups revealed significant differences only for miR-5193.
Several studies have identified a reduction in the miR-214 level in HCC tissues and cell lines (29), but none of the previous studies evaluated the plasma levels of this molecule as a potential biomarker for HCC. MiR-214 is thought to induce its effects by targeting essential molecules in the cell cycle, including maternal embryonic leucine zipper kinase (MELK) mRNA, β-catenin, and zeste homolog 2 (EZH2) (30). MiR-214 might also suppress the development of HCC by targeting E2F3 (31). The bioinformatics predictions in the current study indicated that miR-214 might directly target the HBx mRNA of the virus. Therefore, HCC progression may be prevented by the expression of miR-214 in cancer tissues through decreases in the expression of HBx protein.
The bioinformatics predictions presented here also indicated that miR-34a could target the transcript of the Notch1 gene, as the plasma level of this miRNA was decreased in patients with HCC. The decreased expression of miR-34a has also been reported in the tumorigenesis and progression of many types of cancer, including HCC (32, 33). MiR-34a was reported as a biomarker for predicting the risk of bone metastasis in HCC patients (34). More recently, the downregulation of miR-34a has been associated with disease progression of a poor-prognosis type of pancreatic ductal adenocarcinoma (PDAC) that was resistant to chemotherapy (35). The same study also implied that miR-34a exerted its effect through the inhibition of the Notch1-related signaling pathways.
On the other hand, miR-6510 is a newly identified microRNA for which no biological role has yet been described in the literature. The present study is the first report of the biological function of this molecule. The plasma level of this molecule was significantly reduced in patients suffering from HCC when compared to healthy individuals and patients with chronic HBV infection (-4.23 and -2.48 folds, respectively). Nonetheless, similar to the observations for miR-214 and miR-34a, the plasma level of miR-6510 was not significantly different between healthy people and patients with chronic hepatitis B. These findings confirm that the observed downregulations are related to HCC and not to chronic infection with hepatitis B virus.
In the present study, the most marked aberrant expression was observed for miR-5193 (Figure 1). To our knowledge, there is only one other study in the literature regarding this molecule, reporting that miR-5193 targets HBV transcripts in all HBV genotypes and that this miRNA has a role in HBV replication (36). This information confirms our bioinformatics prediction regarding miR-5193. The RT-qPCR results in the present study showed that the normalized fold changes for miR-5192, when compared to healthy individuals and patients with chronic HBV, were -18.85 and -5.7, respectively. Moreover, when compared to the levels in healthy individuals, this molecule was downregulated in patients with chronic hepatitis B (-3.28 folds). This suggests that this molecule may have an association with HCC through a direct role in HBV persistence.
The biggest challenge in the current study was the small sample size. However, since the prevalence of HDV in Iran is low and it is found lower than 1% in chronic HBV patients and lower than 5% in cirrhotic HBV patients (37), we believe that the HCC group was not populated by individuals with HDV infection and thus, the small sample size in the current study did not influence the obtained results.
The ROC curve analyses were also performed to assess the possibility of using plasma levels of these four miRNAs as diagnostic biomarkers for detecting HCC. In the first calculation, the microRNA levels were compared between HCC patients and healthy individuals. The values for miR-34a and miR-6510 were reasonable, but miR-5193 showed the greatest AUC, sensitivity, and specificity as 96%, 100%, and 0.993, respectively, at a cutoff of 0.266 (Figure 2). Therefore, differences in the expression of miR-5193 could accurately distinguish healthy individuals from patients with HCC, with excellent sensitivity and specificity.
The second ROC analysis compared the plasma levels of the miRNAs between the HCC and chronic HBV groups. The best values were again obtained for miR-5193, which showed sensitivity, specificity, and AUC of 79.8%, 82%, and 0.817, respectively, at a cutoff of 0.612. The values obtained for the other miRNAs examined were not discriminatory between HCC and HBV patients.
5.1. Conclusions
In conclusion, the results of the current study showed that the four predicted miRNAs were downregulated in the plasma of patients with HCC and thus, they may have roles in HCC development and progression. Of the miRNAs examined, miR-5193 appears to be a particularly promising candidate to serve as a novel non-invasive biomarker for the detection of HBV-induced HCC.