Abstract
Context:
The number of quinolone-resistant Vibrio cholerae strains reported in various clinical studies is growing steadily since the 1990s. Important biological conditions such as alterations of DNA topoisomerases, perturbations in regard to drug entry and efflux and plasmids transferability between bacteria, are conditions that lead to the acquisition of certain power to resist quinolones during therapy. The current article aimed to review the evolution of quinolone-resistant strains of V. cholerae, the basis for quinolone mode of action against DNA topoisomerases and the recent advances in the understanding of the mechanisms by which V. cholerae strains are resistant to quinolones, including chromosomal mediated resistance involving mutations in topoisomerases, chromosomal mediated resistance resulted in response to reduced expression and/or overexpression of proteins synthesis for porins and cellular efflux pumps, respectively and plasmid mediated resistance.Evidence Acquisition:
This systematic review utilized the research results of clinical trials.gov database, the cochrane database of systematic reviews, Medline, PubMed, Google and other databases to select about 67 publications on cholera and fluoroquinolones resistance mechanisms in V. cholerae. Relevant publications were reviewed; findings were synthesized using a narrative method to illustrate how the treatment of cholera with quinolones as adjunct is evolved.Results:
Through the thorough literature review, it was found that despite the progress made in the development of effective fluoroquinolones against various enteric pathogens including V. cholerae, new concerns emerged which include the constant reports on reduced susceptibility of these drugs against V. cholerae strains endorsed with the emergence of various mechanisms of resistance.Conclusions:
The current systematic review emphasized on the importance of surveying the emergence of V. cholerae resistance to quinolones that may occur during adjunct treatment of cholera and also the importance of determining the genetic basis of the resistance mechanisms for future development of more promising quinolones.Keywords
1. Context
Rehydration therapy by appropriate oral or intravenous solutions is strongly advised during the treatment of cholera, a gastroenteritis disease with serious public health implications. Various international organizations engaged in promoting and implementing subjects relevant to public health infectious diseases recommend antimicrobial drug treatment for severely dehydrated patients with suspected cholera. Reduced duration of diarrhea and volume of stool and reduced duration of vibrio shedding due to adjunct treatment with antibiotic were reported in a recent Cochrane meta-analysis of several clinical trials conducted on a large sample of patients (1). Amongst various groups of antimicrobials, fluoroquinolones encountered significant emergence to treat gastroenteritis diseases. Evidence for various successes of clinical experience with quinolones in cholera and other enteric infections are reported in various international journals (2). A treatment network with quinolones elucidating various comparisons and the number of trials examining each comparison is presented in Figure 1. Ciprofloxacin, norfloxacin and ofloxacin were compared to placebo/no treatment (comparison not shown in Figure 1).
Treatment network implicating quinolones; different comparisons included studies which compared one quinolone vs another quinolone are described in this figure.
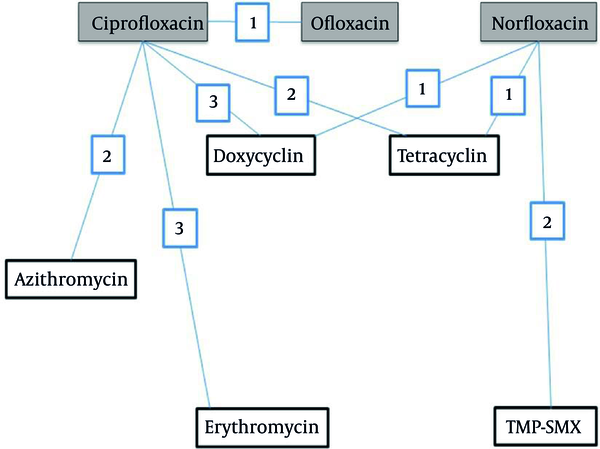
Utility of quinolone as adjunct in the treatment of cholera was first confirmed by norfloxacin as reported in studies first conducted in Calcutta, India (3), then in Lima, Peru (4), in Thailand (5), and in Salta, Argentina (6) respectively. Ciprofloxacin is another important quinolone with confirmed utility against gastroenteritis such as cholera and recorded as the most extensively studied amongst others. Excellent results with this fluoroquinolones was reported in an open, non-controlled study conducted by Doganci et al. (7), in two large randomized double-blind controlled studies conducted in Peru and Bangladesh (8), and in a large clinical trial conducted in Bangladesh (9). Results of these important studies clearly showed that quinolones were highly effective in all clinical parameters.
However, in recent years many studies from Africa, Asia and America reported the emergence of quinolone-resistant strains of V. cholerae (10, 11). Chander et al., and Saha et al., in different studies including 277 and 195 isolates of V. cholerae from Chandigarh (India) and Bangladesh respectively, both reported the emergence of ciprofloxacin-resistant strains of V. cholerae (12-14). In a study including strains of V. cholerae species isolated from 61 cases with gastroenteritis admitted to the gastroenteritis wards of Karnataka, Hubli (South India) and hospitalized for one to seven days during the cholera epidemic of 2002, Krishna et al. reported norfloxacin and ciprofloxacin resistance with the prevalence of 12.5% (15). A warning record on progressive increases in ciprofloxacin and norfloxacin resistance among V. cholerae O1 strains with the highest prevalence of 38.8% in 1999 and 25% in 2000, respectively was demonstrated by Garg et al., in a study on patients with cholera admitted to the Infectious Diseases Hospital, Calcutta, India (16). Mercy et al., in a study on V. cholerae O1 species isolated from different regions of Kenya from 2007 to 2010 reported resistance against nalidixic acid and ciprofloxacin with the prevalence of 89% and 2.3%, respectively (17). In a study aimed to determine the antibiotic susceptibility patterns of 118 strains of V. cholerae O1 isolated in 2010 at Jawaharlal Nehru institute of post graduate medical education and research, Puducherry, India, Mandal et al., reported resistance to ciprofloxacin with a modest occurrence of 4.2% (18). While a study by Eibach et al., reported that the increase in ciprofloxacin resistance reached about 98% in 2014 (19) (Table 1).
Vibrio cholerae O1 Species Isolates From Different Cholera Outbreaks and the Prevalence of Resistance to Quinolones
| Year | Number of Isolates | Number of Resistant Strains (%) | |||
|---|---|---|---|---|---|
| Nalidixic Acid | Ciprofloxacin | Norfloxacin | Ref. | ||
| 1989 | 49 | 3 (6.1) | ND | ND | (16) |
| 1990 | 59 | 1 (1.7) | ND | ND | (16) |
| 1991 | 30 | ND | ND | (16) | |
| 1992 | 26 | 2 (7.7) | (16) | ||
| 1993 | 20 | 1 (5) | (16) | ||
| 1994 | 74 | 73 (98.6) | (16) | ||
| 1995 | 84 | 82 (97.6) | 2 (2.4) | 3(3.6) | (16) |
| 1996 | 69 | 68 (98.5) | 4 (5.8) | (16) | |
| 1997 | 53 | 50 (94.3) | 10 (18.9) | 4 (7.5) | (16) |
| 1998 | 201 | 197 (98) | 20 (10) | 1 (0.5) | (16) |
| 1999 | 49 | 49 (100) | 19 (38.8) | 8 (16.3) | (16) |
| 2000 | 16 | 12 (75) | 3 (18.7) | 4 (25) | (16) |
| 2007-2010 | 44 | 39 (89) | 1 (2.3) | (17) | |
| 2010 | 118 | ND | 5 (4.2) | ND | (18) |
| 2014 | 92 | 62 (100) | 61 (98.4) | (19) | |
2. Evidence Acquisition
Results of the search in clinical trials.gov database and the cochrane database of systematic reviews by John Wiley & Sons, Ltd. were synthesized to illustrate and/or relate the reported clinical success and/or limits of quinolones as adjuncts to treat cholera. Medline, PubMed, Google and other databases were also used to select about 80 publications on cholera and quinolones resistant mechanisms in V. cholera to evaluate how the treatment of cholera with these drugs is evolved regarding the various quinolone-resistant V. cholerae strains.
3. Results
3.1. Quinolone Mechanisms of Activity
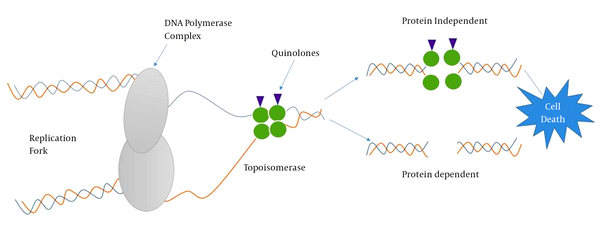
Understanding antibiotic resistance mechanisms requires an understanding of where antibiotics exert their effect. Cellular events such as DNA replication and mRNA transcription that take place during cell cycle or cell-division cycle require the intervention of topoisomerases such as DNA gyrase (type II topoisomerase) recognized to participate in relieving some of the stress during DNA synthesis (20). Thus, chemists and pharmacists involved in the synthesis of quinolones do exploit these conditions to target DNA-topoisomerase complexes (21, 22). Quinolones are bactericidal compounds that inhibit bacterial DNA from unwinding and duplicating through inhibition of the topoisomerase II ligase domain. This interference consequently leads to transitory DNA fragmentation and by catalyzing the passage of DNA segments within the cuts before closing them and though leading to rapid cell death (Figure 2). DNA gyrase, also referred as topoisomerase II, consists of two polypeptide subunits (GyrA and GyrB) with the function to regulate links via relaxing supercoils and by introducing both positive and negative supercoils between DNA double strands. The A subunit (encoded by gyrA gene) carries out binding of DNA, B subunit mediates the introduction of negative supercoils, and then A subunit participates in the reunion of DNA strands (23, 24). Topoisomerase IV, similar to topoisomerase II, consists of two polypeptide subunits (ParC and ParE) with the role to participate in separating new replicates of chromosomes following the replication process. Quinolones binds to the A subunit of topoisomerase II and though interfering in various steps involved in the DNA replication process.
Quinolone-Target Interactions and Associated Cell Death Mechanisms
3.2. Chromosomal Mediated Resistance Involving Mutations in Topoisomerase
In many bacteria, such as V. cholerae, quinolones resistance is a chromosomal mediated resistance involving mutations in the quinolone resistance-determining regions (QRDRs) of topoisomerases genes gyrA and parC. Quinolone resistance in V. cholerae species is mainly observed in strains harboring mutations in gyrA and parC genes of topoisomerase II and IV respectively (25, 26). Substitutions of single amino acid (Ser-83; Ile in gyrA and Ser-85; Leu in parC) were observed in genes of topoisomerase II and IV of V. cholerae O1 with marked reduced susceptibility to ciprofloxacin (27).
3.3. Efflux Resistance Mechanisms
The efflux system pumps out a variety of structurally and functionally distinct antimicrobials including quinolones from bacterial cells. To date, advancement in biomedicine research oriented through the understanding of the biochemistry of Gram-negative bacteria cell wall and the mechanisms involved in resisting antimicrobials such as quinolones significantly contributed to the discovery and characterization of various multidrug and drug-specific efflux pumps from bacteria.
Vibrio cholerae efflux pumps are categorized into two major families, such as the ATP-binding cassette (ABC) superfamily (28), and the proton-motive force (PMF) pump families. The PMF sub-families include the major facilitator superfamily (MFS) (29), the multidrug and toxic compound extrusion (MATE) family (30), the small multidrug resistance (SMR) family (31) and the resistance-nodulation-division (RND) superfamily (32-34). In V. cholerae, six putative genes from MATE family of efflux pumps are reported. Involvement of genes such as VcmB, VcmD, VcmH, VcmN, VcmA and VcrM (35) with energy dependent efflux toward fluoroquinolones is demonstrated in Escherichia coli and V. cholerae background. Recently, homologue of NorM in Vibrio parahaemolyticus (36) reported to mediate resistance to hydrophilic fluoroquinolones, aminoglycosides and norfloxacin was discovered in the genome of V. cholerae O1 El Tor N16961 (37). Among the V. cholerae efflux systems, MFS transporters and the PMF-dependent efflux were reported in separate studies to confer resistance to nalidixic acid (38) and various quinolones (39) in clinical strains of V. cholerae. RND efflux systems were encoded by six operons. Four operons (VexAB, VexCD, VexGH and VexIJK) out of six were reported to confer resistance to antimicrobials in V. cholerae (40-42). VexAB was associated with resistance to several antimicrobials determined by mutational analysis (40). In a recent study conducted by Jun O et al., inherently reduced expression of gene vca0421 in V. cholerae O1 was demonstrated to confer resistance to fluoroquinolones in V. cholerae O1 (43-47).
3.4. Plasmid Mediated Quinolone Resistance
Plasmids carrying genes that encode drug resistance determinants spread very fast in mixed bacteria communities. However, the wild strains of V. cholerae are mostly reported as poor hosts for plasmids carrying genes. The frequency of plasmid carriage in clinical strains of V. cholerae O1 was less (2%) compared with clinical non-O1 strains (25%) as reported elsewhere. Most of the clinical strains of V. cholerae O1 isolated from different geographical regions lacked the plasmids (48, 49), and when present, the size and functions of plasmids were not uniform. A study reported some of the naturally occurring conjugative and non-conjugative plasmids harboring drug resistance determinants in environmental and clinical isolates of V. cholerae encoding genes inducing various antibiotics resistance.
Conjugative plasmids evolved to increase the relative fitness of bacteria had implications both for maintenance and spread of plasmids. Transfer of multiple-drug-resistance from V. cholerae to E. coli (K12) was demonstrated in-vitro with conjugative plasmids. Transfer of multiple-drug-resistant (MDR) plasmids of group InC through conjugation with other bacteria was reported in a study conducted by Glass et al. This plasmid conferred resistance to a number of antibiotics including quinolones (50). MDR V. cholerae species isolated in Kenya harbored a 100 MDa plasmid of group IncC (51). In Algeria, the IncJ plasmids (IncJ incompatibility group) containing antibiotic resistance marker was identified for the first time in V. cholerae El Tor (52).
An emerging clinical concern that causes reduced susceptibility to quinolones was experienced with plasmids identified to carry quinolone resistant genes (53).Plasmid-quinolone resistant genes include intrinsic or qnr genes (22, 54, 55), reported to protect topoisomerases from quinolones in bacteria (56). Modest findings reported in a study on V. cholerae isolates in Brazil described qnrVC1, a homolog to qnr. Limitations regarding this study include lack of molecular mechanisms evidencing that qnrVC1 could be linked to reduced susceptibility of ciprofloxacin in V. cholerae (57). However, Hong Bin Kim et al., in a recent study on V. cholerae species isolated from Bangladesh, described a new qnr homolog, qnrVC3 and a large number of mutations in topoisomerase target enzymes that confer higher levels of transferable quinolone resistance (58, 59).
4. Conclusions
The application of antibiotics as adjunct to treat patients with cholera infection is encouraged by various international health organizations such as world health organization (WHO) and Medecin Sans Frontiere (MSF). Quinolones are one of the most important classes of antibiotics available to treat gastroenteritis including cholera. The emergence of multidrug or specific drug resistant V. cholerae strains significantly affected the clinical contribution of these drugs. To date, various studies reported comprehensive descriptions of various molecular mechanisms and mutations in topoisomerases enzymes involved in or causing drug resistance, respectively. Quinolones are in the clinics to treat cholera for several decades, but knowledge about the mechanisms of interaction between quinolone and its specific targets (i e, topoisomerases enzymes) and how mutations cause resistance is only in progress but not conclusive. Since the emergence of quinolones-resistant V. cholerae may significantly influence the strategies of controlling cholera, locally and regionally based surveillance information on antimicrobial susceptibility of V. cholerae should give importance to this aspect. Hopefully, this review can be used as a resource document to direct the development of new promising generation of quinolones to treat cholera based on better understanding of the molecular mechanisms of action of these synthetic drugs.
Acknowledgements
References
-
1.
Leibovici-Weissman Y, Neuberger A, Bitterman R, Sinclair D, Salam MA, Paul M. Antimicrobial drugs for treating cholera. Cochrane Database Syst Rev. 2014;(6):CD008625. [PubMed ID: 24944120]. https://doi.org/10.1002/14651858.CD008625.pub2.
-
2.
Seas C, Gotuzzo E. Vibrio cholerae (Cholera). USA: www.antimicrobe.org; 2014.
-
3.
Bhattacharya SK, Bhattacharya MK, Dutta P, Dutta D, De SP, Sikdar SN, et al. Double-blind, randomized, controlled clinical trial of norfloxacin for cholera. Antimicrob Agents Chemother. 1990;34(5):939-40. [PubMed ID: 2193628].
-
4.
Gotuzzo E, Cieza J, Estremadoyro L, Seas C. Cholera. Lessons from the epidemic in Peru. Infect Dis Clin North Am. 1994;8(1):183-205. [PubMed ID: 7980768].
-
5.
Moolasart P, Eampokalap B, Supaswadikul S. Comparison of the efficacy of tetracycline and norfloxacin in the treatment of acute severe watery diarrhea. Southeast Asian J Trop Med Public Health. 1998;29(1):108-11. [PubMed ID: 9740281].
-
6.
Seijo AC, Fernandez M, Campos C, Cernigoi BE, Nogueras M, San Juan J, et al. Norfloxacin monodose use in patients with cholera in Salta Argentina. Rev Inst Med Trop Sao Paulo. 1996;38(3):217-9. [PubMed ID: 9163987].
-
7.
Doganci L, Gun H, Baysallar M, Albay A, Cinar E, Haznedaroglu T. Short-term quinolones for successful eradication of multiply resistant Vibrio cholerae in adult patients. Scand J Infect Dis. 1995;27(4):425-6. [PubMed ID: 8658086].
-
8.
Gotuzzo E, Seas C, Echevarria J, Carrillo C, Mostorino R, Ruiz R. Ciprofloxacin for the treatment of cholera: a randomized, double-blind, controlled clinical trial of a single daily dose in Peruvian adults. Clin Infect Dis. 1995;20(6):1485-90. [PubMed ID: 7548496].
-
9.
Khan W, Bennish ML, Seas C, Khan EH, Ronan A, Dhar U, et al. Randomised controlled comparison of single-dose ciprofloxacin and doxycycline for cholera caused by Vibrio cholerae O1 or O139. The Lancet. 1996;348(9023):296-300. https://doi.org/10.1016/s0140-6736(96)01180-4.
-
10.
Jesudason MV, John TJ. Transferable trimethoprim resistance of Vibrio cholerae O1 encountered in southern India. Trans R Soc Trop Med Hyg. 1990;84(1):136-7. [PubMed ID: 2345916].
-
11.
Faruque AS, Alam K, Malek MA, Khan MG, Ahmed S, Saha D, et al. Emergence of multidrug-resistant strain of Vibrio cholerae O1 in Bangladesh and reversal of their susceptibility to tetracycline after two years. J Health Popul Nutr. 2007;25(2):241-3. [PubMed ID: 17985827].
-
12.
Bauer AW, Kirby WM, Sherris JC, Turck M. Antibiotic susceptibility testing by a standardized single disk method. Am J Clin Pathol. 1966;45(4):493-6. [PubMed ID: 5325707].
-
13.
Chander J, Kaistha N, Gupta V, Mehta M, Singla N, Deep A, et al. Epidemiology & antibiograms of Vibrio cholerae isolates from a tertiary care hospital in Chandigarh, north India. Indian J Med Res. 2009;129(5):613-7. [PubMed ID: 19675394].
-
14.
Saha D, Karim MM, Khan WA, Ahmed S, Salam MA, Bennish ML. Single-dose azithromycin for the treatment of cholera in adults. N Engl J Med. 2006;354(23):2452-62. [PubMed ID: 16760445]. https://doi.org/10.1056/NEJMoa054493.
-
15.
Krishna BV, Patil AB, Chandrasekhar MR. Fluoroquinolone-resistant Vibrio cholerae isolated during a cholera outbreak in India. Trans R Soc Trop Med Hyg. 2006;100(3):224-6. [PubMed ID: 16246383]. https://doi.org/10.1016/j.trstmh.2005.07.007.
-
16.
Garg P, Sinha S, Chakraborty R, Bhattacharya SK, Nair GB, Ramamurthy T, et al. Emergence of fluoroquinolone-resistant strains of Vibrio cholerae O1 biotype El Tor among hospitalized patients with cholera in Calcutta, India. Antimicrob Agents Chemother. 2001;45(5):1605-6. [PubMed ID: 11372642].
-
17.
Mercy N, Mohamed AA, Zipporah N, Chowdhury G, Pazhani GP, Ramamurthy T, et al. Phenotypic and genetic characterization of Vibrio cholerae O1 isolated from various regions of Kenya between 2007 and 2010. Pan Afr Med J. 2014;19:8. [PubMed ID: 25584121]. https://doi.org/10.11604/pamj.2014.19.8.2496.
-
18.
Mandal J, Dinoop KP, Parija SC. Increasing antimicrobial resistance of Vibrio cholerae OI biotype E1 tor strains isolated in a tertiary-care centre in India. J Health Popul Nutr. 2012;30(1):12-6. [PubMed ID: 22524114].
-
19.
Eibach D, Herrera-Leon S, Gil H, Hogan B, Ehlkes L, Adjabeng M, et al. Molecular Epidemiology and Antibiotic Susceptibility of Vibrio cholerae Associated with a Large Cholera Outbreak in Ghana in 2014. PLoS Negl Trop Dis. 2016;10(5):4751. [PubMed ID: 27232338]. https://doi.org/10.1371/journal.pntd.0004751.
-
20.
Espeli O, Marians KJ. Untangling intracellular DNA topology. Mol Microbiol. 2004;52(4):925-31. [PubMed ID: 15130115]. https://doi.org/10.1111/j.1365-2958.2004.04047.x.
-
21.
Drlica K, Malik M, Kerns RJ, Zhao X. Quinolone-mediated bacterial death. Antimicrob Agents Chemother. 2008;52(2):385-92. [PubMed ID: 17724149]. https://doi.org/10.1128/AAC.01617-06.
-
22.
Sugino A, Peebles CL, Kreuzer KN, Cozzarelli NR. Mechanism of action of nalidixic acid: purification of Escherichia coli nalA gene product and its relationship to DNA gyrase and a novel nicking-closing enzyme. Proc Natl Acad Sci U S A. 1977;74(11):4767-71. [PubMed ID: 200930].
-
23.
Willmore E, de Caux S, Sunter NJ, Tilby MJ, Jackson GH, Austin CA, et al. A novel DNA-dependent protein kinase inhibitor, NU7026, potentiates the cytotoxicity of topoisomerase II poisons used in the treatment of leukemia. Blood. 2004;103(12):4659-65. [PubMed ID: 15010369]. https://doi.org/10.1182/blood-2003-07-2527.
-
24.
Chen CR, Malik M, Snyder M, Drlica K. DNA gyrase and topoisomerase IV on the bacterial chromosome: quinolone-induced DNA cleavage. J Mol Biol. 1996;258(4):627-37. [PubMed ID: 8636997]. https://doi.org/10.1006/jmbi.1996.0274.
-
25.
Everett MJ, Jin YF, Ricci V, Piddock LJ. Contributions of individual mechanisms to fluoroquinolone resistance in 36 Escherichia coli strains isolated from humans and animals. Antimicrob Agents Chemother. 1996;40(10):2380-6. [PubMed ID: 8891148].
-
26.
Vila J, Ruiz J, Marco F, Barcelo A, Goni P, Giralt E, et al. Association between double mutation in gyrA gene of ciprofloxacin-resistant clinical isolates of Escherichia coli and MICs. Antimicrob Agents Chemother. 1994;38(10):2477-9. [PubMed ID: 7840592].
-
27.
Quilici ML, Massenet D, Gake B, Bwalki B, Olson DM. Vibrio cholerae O1 variant with reduced susceptibility to ciprofloxacin, Western Africa. Emerg Infect Dis. 2010;16(11):1804-5. [PubMed ID: 21029554]. https://doi.org/10.3201/eid1611.100568.
-
28.
Lubelski J, Konings WN, Driessen AJ. Distribution and physiology of ABC-type transporters contributing to multidrug resistance in bacteria. Microbiol Mol Biol Rev. 2007;71(3):463-76. [PubMed ID: 17804667]. https://doi.org/10.1128/MMBR.00001-07.
-
29.
Pao SS, Paulsen IT, Saier MH. Major facilitator superfamily. Microbiol Mol Biol Rev. 1998;62(1):1-34. [PubMed ID: 9529885].
-
30.
Kuroda T, Tsuchiya T. Multidrug efflux transporters in the MATE family. Biochim Biophys Acta. 2009;1794(5):763-8. [PubMed ID: 19100867]. https://doi.org/10.1016/j.bbapap.2008.11.012.
-
31.
Jack DL, Yang NM, Saier MH. The drug/metabolite transporter superfamily. Eur J Biochem. 2001;268(13):3620-39. [PubMed ID: 11432728].
-
32.
Nikaido H, Takatsuka Y. Mechanisms of RND multidrug efflux pumps. Biochim Biophys Acta. 2009;1794(5):769-81. [PubMed ID: 19026770]. https://doi.org/10.1016/j.bbapap.2008.10.004.
-
33.
Paulsen IT, Brown MH, Skurray RA. Proton-dependent multidrug efflux systems. Microbiol Rev. 1996;60(4):575-608. [PubMed ID: 8987357].
-
34.
Putman M, van Veen HW, Konings WN. Molecular properties of bacterial multidrug transporters. Microbiol Mol Biol Rev. 2000;64(4):672-93. [PubMed ID: 11104814].
-
35.
Begum A, Rahman MM, Ogawa W, Mizushima T, Kuroda T, Tsuchiya T. Gene cloning and characterization of four MATE family multidrug efflux pumps from Vibrio cholerae non-O1. Microbiol Immunol. 2005;49(11):949-57. [PubMed ID: 16301805].
-
36.
Heidelberg JF, Eisen JA, Nelson WC, Clayton RA, Gwinn ML, Dodson RJ, et al. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature. 2000;406(6795):477-83. [PubMed ID: 10952301]. https://doi.org/10.1038/35020000.
-
37.
Morita Y, Kodama K, Shiota S, Mine T, Kataoka A, Mizushima T, et al. NorM, a putative multidrug efflux protein, of Vibrio parahaemolyticus and its homolog in Escherichia coli. Antimicrob Agents Chemother. 1998;42(7):1778-82. [PubMed ID: 9661020].
-
38.
Colmer JA, Fralick JA, Hamood AN. Isolation and characterization of a putative multidrug resistance pump from Vibrio cholerae. Mol Microbiol. 1998;27(1):63-72. [PubMed ID: 9466256].
-
39.
Baranwal S, Dey K, Ramamurthy T, Nair GB, Kundu M. Role of active efflux in association with target gene mutations in fluoroquinolone resistance in clinical isolates of Vibrio cholerae. Antimicrob Agents Chemother. 2002;46(8):2676-8. [PubMed ID: 12121955].
-
40.
Bina JE, Provenzano D, Wang C, Bina XR, Mekalanos JJ. Characterization of the Vibrio cholerae vexAB and vexCD efflux systems. Arch Microbiol. 2006;186(3):171-81. [PubMed ID: 16804679]. https://doi.org/10.1007/s00203-006-0133-5.
-
41.
Bina XR, Provenzano D, Nguyen N, Bina JE. Vibrio cholerae RND family efflux systems are required for antimicrobial resistance, optimal virulence factor production, and colonization of the infant mouse small intestine. Infect Immun. 2008;76(8):3595-605. [PubMed ID: 18490456]. https://doi.org/10.1128/IAI.01620-07.
-
42.
Van Bambeke F, Michot JM, Tulkens PM. Antibiotic efflux pumps in eukaryotic cells: occurrence and impact on antibiotic cellular pharmacokinetics, pharmacodynamics and toxicodynamics. J Antimicrob Chemother. 2003;51(5):1067-77. [PubMed ID: 12697641]. https://doi.org/10.1093/jac/dkg225.
-
43.
Okuda J, Hayashi N, Wakahara Y, Gotoh N. Reduced expression of the vca0421 gene of Vibrio cholerae O1 results in innate resistance to ciprofloxacin. Antimicrob Agents Chemother. 2010;54(11):4917-9. [PubMed ID: 20805389]. https://doi.org/10.1128/AAC.01652-09.
-
44.
Li XZ, Nikaido H. Efflux-mediated drug resistance in bacteria. Drugs. 2004;64(2):159-204. [PubMed ID: 14717618].
-
45.
Chen J, Morita Y, Huda MN, Kuroda T, Mizushima T, Tsuchiya T. VmrA, a member of a novel class of Na(+)-coupled multidrug efflux pumps from Vibrio parahaemolyticus. J Bacteriol. 2002;184(2):572-6. [PubMed ID: 11751837].
-
46.
Huda N, Lee EW, Chen J, Morita Y, Kuroda T, Mizushima T, et al. Molecular cloning and characterization of an ABC multidrug efflux pump, VcaM, in Non-O1 Vibrio cholerae. Antimicrob Agents Chemother. 2003;47(8):2413-7. [PubMed ID: 12878498].
-
47.
Woolley RC, Vediyappan G, Anderson M, Lackey M, Ramasubramanian B, Jiangping B, et al. Characterization of the Vibrio cholerae vceCAB multiple-drug resistance efflux operon in Escherichia coli. J Bacteriol. 2005;187(15):5500-3. [PubMed ID: 16030246]. https://doi.org/10.1128/JB.187.15.5500-5503.2005.
-
48.
Bag PK, Maiti S, Sharma C, Ghosh A, Basu A, Mitra R, et al. Rapid spread of the new clone of Vibrio cholerae O1 biotype El Tor in cholera endemic areas in India. Epidemiol Infect. 1998;121(2):245-51. [PubMed ID: 9825773].
-
49.
Clark CA, Purins L, Kaewrakon P, Manning PA. VCR repetitive sequence elements in the Vibrio cholerae chromosome constitute a mega-integron. Mol Microbiol. 1997;26(5):1137-8. [PubMed ID: 9426148].
-
50.
Glass RI, Huq MI, Lee JV, Threlfall EJ, Khan MR, Alim AR, et al. Plasmid-borne multiple drug resistance in Vibrio cholerae serogroup O1, biotype El Tor: evidence for a point-source outbreak in Bangladesh. J Infect Dis. 1983;147(2):204-9. [PubMed ID: 6827137].
-
51.
Finch MJ, Morris JG, Kaviti J, Kagwanja W, Levine MM. Epidemiology of antimicrobial resistant cholera in Kenya and East Africa. Am J Trop Med Hyg. 1988;39(5):484-90. [PubMed ID: 3195696].
-
52.
Korichi MN, Belhocine S, Rahal K. [Inc J plasmids identified for the first time in Vibrio cholerae El Tor]. Med Trop (Mars). 1997;57(3):249-52. [PubMed ID: 9513150].
-
53.
Martinez-Freijo P, Fluit AC, Schmitz FJ, Grek VS, Verhoef J, Jones ME. Class I integrons in Gram-negative isolates from different European hospitals and association with decreased susceptibility to multiple antibiotic compounds. J Antimicrob Chemother. 1998;42(6):689-96. [PubMed ID: 10052890].
-
54.
Robicsek A, Jacoby GA, Hooper DC. The worldwide emergence of plasmid-mediated quinolone resistance. Lancet Infect Dis. 2006;6(10):629-40. [PubMed ID: 17008172]. https://doi.org/10.1016/S1473-3099(06)70599-0.
-
55.
Robicsek A, Strahilevitz J, Jacoby GA, Macielag M, Abbanat D, Park CH, et al. Fluoroquinolone-modifying enzyme: a new adaptation of a common aminoglycoside acetyltransferase. Nat Med. 2006;12(1):83-8. [PubMed ID: 16369542]. https://doi.org/10.1038/nm1347.
-
56.
Tran JH, Jacoby GA, Hooper DC. Interaction of the plasmid-encoded quinolone resistance protein Qnr with Escherichia coli DNA gyrase. Antimicrob Agents Chemother. 2005;49(1):118-25. [PubMed ID: 15616284]. https://doi.org/10.1128/AAC.49.1.118-125.2005.
-
57.
Fonseca EL, Dos Santos Freitas F, Vieira VV, Vicente AC. New qnr gene cassettes associated with superintegron repeats in Vibrio cholerae O1. Emerg Infect Dis. 2008;14(7):1129-31. [PubMed ID: 18598639]. https://doi.org/10.3201/eid1407.080132.
-
58.
Burrus V, Marrero J, Waldor MK. The current ICE age: biology and evolution of SXT-related integrating conjugative elements. Plasmid. 2006;55(3):173-83. [PubMed ID: 16530834]. https://doi.org/10.1016/j.plasmid.2006.01.001.
-
59.
Kim HB, Wang M, Ahmed S, Park CH, LaRocque RC, Faruque ASG, et al. Transferable quinolone resistance in vibrio cholerae. Antimicrob Agents Chemother. 2009;54(2):799-803. https://doi.org/10.1128/aac.01045-09.