Abstract
Keywords
Experimental Epitope OMP31 IL-2 Expression Brucellosis Escherichia coli
1. Background
There are different genus of Brucella bacteria, which cause considerable economic loss in developing countries. Infection by B. melitensis (mainly cause ovine brucellosis) is known as one of the most frequent and extensive zoonotic worldwide diseases, therefore, many researches have focused on fighting with this disease (1). The outer membrane proteins (OMPs) of these bacteria play a key role in immunity responses; therefore, they can be applied to design recombinant vaccines. OMP31 is known as the most immune-dominant, protective, and exposed antigen of smooth B. malitensis (2). Once the host encounters the pathogens, the antibodies are secreted through detection of specific areas of the antigen that is called “epitope” (3). Previous researches identified an epitope by anti-Omp31 MAb A59/10F09/G10, which consisted of 36 amino acids of Omp31 protein, which represents the most hydrophilic part of the protein (4). Cassataro et al demonstrated that a peptide consisting of 27 amino acid derived from OPM31 protein (OMP3148-74) as a protective epitope can induce immune response against B. melitensis similar to the response from whole protein of OMP31 (2). Using subunit vaccines in immunological tests, based on proprietary T-cell or B-cell epitopes, is safer and more efficient compared to using attenuated and killed vaccines (5). Small peptides have a low half-life due to their low weight and being fast removed from the kidney filtration system, consequently, these peptides need to be fused with a heavier molecule for better activity (6). The goal of vaccination is to induce protective immunity, in some vaccines this parameter can be enhanced by applying some molecular adjuvants. In recent years, cytokines, as molecular adjuvants, are widely being employed to fortify immune responses (7). IL-2, which is mainly produced by CD4+ T cells, is a lymphoproliferative cytokine, which triggers T-Cell differentiation, proliferation, and cellular immune responses, which can be an appropriate candidate as molecular adjuvant (6). Nowadays, fusion proteins are being used as biomolecules with several functions such as prolonging their plasma half-lives, increasing their therapeutic efficacy and drug targeting (e.g. toxin or cytokine) (8). Direct fusion of domains without any linker can cause unwanted results including: misfolding, low yield production, and incomplete biologic activity of fusion proteins (9). Therefore, applying proper linker to fuse protein domains is of importance. For example, helical linker sequences (EAAAK) n, where n can vary from 3-6. Of all the non-helical linkers, we can mention (XP) n, in which x represents Ala, Lys, or Glu amino acids and P represents Pro amino acid. The presence of proline in non-helical linkers increases the linker’s rigidness and separates the domains, which by improving the folding, it can affect protein expression and functionality (8).
2. Objectives
In this study, we have tried to design and express a recombinant chimeric protein (IL2-OMP31 (48-74)3) in prokaryote expression system to investigate its immunity against Brucella melitensis in mice.
3. Methods
3.1. In-Silico Designing and Immunity Characterization
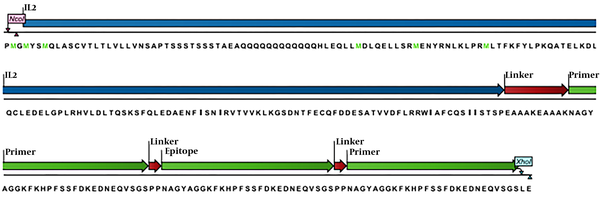
The codon optimized Musmusculus Interleukin-2 (IL-2) cDNA for the expression of Escherichia coli was kindly donated by Biotechnology Laboratory of Animal Science (Ferdowsi University of Mashhad, Iran) in PGH vector (Macrogen, Korea). The coding DNA sequence of three replications of 27 amino acid derived from experimental epitope region of OPM31 (OMP3148-74)3 protein of Brucella melitensis and (EAAAK)2 as a rigid linker bridged to IL-2 (10), were synthesized and inserted in PGH vector (Macrogen, Korea). To determine the antigenicity score of the final desired engineered construction (Figure 1), VaxiJen 2.0 server (http://www.ddg-pharmfac.net/vaxijen/VaxiJen/VaxiJen.html) was employed with a threshold of (0.5).
Schema of amino acid profile of the chimera protein (IL2-3E) which designed by the CLC software; the blue color refers to the IL-2 cytokine amino acids as molecule adjuvant, the green colors are repeats of epitope amino acids and the red colors are the linkers amino acids among domains.
3.2. Secondary and Tertiary Structure Prediction of IL2-OMP31 (48-74)3
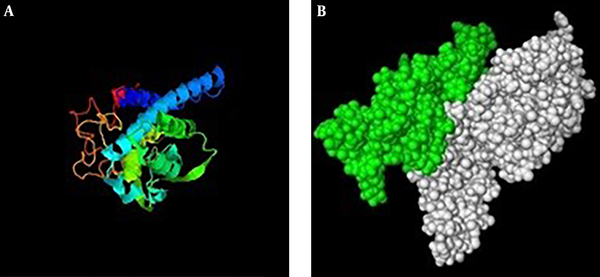
Secondary structure of IL2-OMP31 (48-74)3 recombinant protein (Figure 2) was predicted based on their primary amino acid sequences. In this regard, improved self-optimized prediction method (SOPMA) software (http://npsapbil.ibcp.fr/cgi-bin/npsa_automat.pl?page=/NPSA/npsa_sopma.html) was employed to determine their helices, sheets, turns, and coil structures. Tertiary structures of IL2-OMP31 (48-74)3 protein was predicted by iterative threading assembly refinement (I-TASSER) online server (http://zhanglab.ccmb.med.umich.edu/I-TASSER/) as a hierarchical approach. Finally, PyMOL V1 Viewer software was used to visualize PDB format of the predicted tertiary structure of this protein.
Third structure of chimera protein (IL2-3E); A, predicted structure by I-tasser server. B, PDB format visualizing of the predicted tertiary structure of chimera protein by PyMol software. Green colure demonstrates epitope regions and the whit colure represents IL-2 protein.
3.3. Engineering the Recombinant Construct
Codon optimized murine Interleukin-2 (IL-2) with 519 bp length and experimental epitope region of OMP31 (48-74)3 with rigid linker (3E-L) by 291 bp length sequences were used as templates in PCR reaction. In this regard, IL-2 and 3E-L sequences were amplified using one pair of specific primers, (Table 1). PCR was carried out in a 25 µL reaction volume containing approximately 50 - 100 ng template DNA, 2.5 μL of 10 × PCR buffer, 2 μL dNTPs (2.5 mM), 2 μL MgCl2 (10 mM), 1.5 μL of mix primer (5 pmol/μL), 0.125U/μL of X’Taq DNA polymerase (Takara, Japan), and finally deionized water up to a volume of 25 μL. Thermal cycling was carried out in AB 2720 thermal cycler (Applied Biosystem, USA) using the protocol performed with pre-denaturation at 95°C for 5 minutes, followed by 35 cycles of denaturation at 95°C for 30 seconds, annealing at 56°C and 62°C for 30 seconds for IL-2 and 3E-L respectively, and extension at 72°C for 30 seconds. The final extension step was done at 72°C for 5 minutes. Aliquot of PCR products were analyzed by electrophoresis on 1.5% agarose gel in 1X TBE buffer with a 1kp DNA size marker (Fermentase, Germany), and then virtualized with gel documentation system (BioRad, USA).
The List of Primers Used in This Study
To construct chimeric sequence (IL2-3E), SOE (Splicing by overhang extension) PCR procedure was employed. Previous PCR products (3E-L and IL-2) were used as templates in SOE-PCR reaction without any primers (Figure 1). This reaction was conducted in 25 µL total volume containing approximately 50 - 100 ng of both PCR products, and mixture of PCR ingredients as before. In PCR optimal conditions after denaturation at 94°C for 5 minutes, 15 amplification cycles were performed: denaturation at 94°C for 30 seconds, annealing at 60°C for 30 seconds, extension at 72°C for 45 seconds followed by a final extension at 72°C for 10 minutes. The final phase of SOE-PCR was amplification of target chimera scaffold using the products in previous steps. In this step 5 pmol/μL of IL-2-F and 3E-L-R primers and standard mixture of other components as described before was used with the initial denaturation at 94°C for 5 minutes and followed by 25 cycles of denaturation for 30s at 94°C, 30 seconds at 60°C (annealing step), 30 seconds at 72°C (extension step), and the final extension at 72°C for 5 minutes.
3.4. Gene Cloning and Sub-Cloning
Chimeric fragment (3E-IL2) was purified from 1% agarose gel using Agarose Gel Mini Prep Kit (BioRon, Germany). The purified products were ligated into pTZ57R/T cloning vector by T/A cloning strategy, according to the manufacturer’s instructions (Thermo, USA). Escherichia coli DH5α competent cell preparation and transformation steps were followed as described by Sambrook et al. (11). The recombinant plasmids were transformed into competent E. coli DH5α through heat shock process. The recombinant clone(s) was screened based on their ampicillin resistance and approved by colony PCR (using M13 universal primers) and restriction sites enzyme digestion (XhoI & NcoI).
Then ampicillin-resistant colonies were grown in Luria broth (LB) medium containing 50 µg/mL of ampicillin at 37°C for 16 hours in an incubator. Recombinant plasmids were purified using Plasmid DNA isolation NC Kit (Denazist, Iran) and were confirmed by restriction enzyme digestion.
In order to sub-clone the recombinant construction into pET-22b (+) as an expression vector (Novagen, USA), both pET-22b (+) and pTZ57R/T (containing chimeric fragment ) vectors were enzymatically double-digested by XhoI and NcoI restriction enzymes (Thermo, USA) for 5 hours; final digested products were extracted from 1% agarose gel using Agarose Gel Mini Prep Kit (Thermo, USA), then they went through ligation process according to the manufacturer’s instructions kit (Thermo, USA). Standard techniques for the next steps such as ligation, competent cell preparation and transformation (into competent E. coli BL21 (DE3)) were followed as described previously (11). The recombinant clone (s) were screened based on their ampicillin resistance. The fidelity of E. coli BL21 (DE3) transformants were confirmed by colony-PCR and T7 universal primers and sequencing.
3.5. Expression and Purification of Recombinant Protein
Expression process was performed through culturing positive recombinant BL21 (D3) colonies in 300 mL LB medium (12). The bacteria were harvested by centrifugation (3000 g, 20 minutes, 4°C) and were resuspended in 10 mL of TSE buffer (Tris-HCL 200 mM, pH = 8.0, Sacaros 500 mM, EDTA1 mM,) to extract recombinant protein from periplasmic area. The recombinant protein extraction was followed by incubation for 30 minutes on ice and then this suspension was centrifuged in 16000g for 30 minutes (4°C). The supernatant containing recombinant protein was evaluated on sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) 12%. Protein purification was carried out by chromatography using Ni-agarose (Thermo, USA) according to the manufacturer’s protocol. The quality and identity of the purified rCIL2-3E protein were analyzed by 12% SDS-PAGE.
3.6. Qualitative and Quantitative Evaluation of Recombinant Protein
Western blot process was carried out by electro blotting SDS-PAGE gels onto nitrocellulose membrane. The membrane was dried for 10 minutes in an incubator. Then non-specific sites of blotted nitrocellulose were blocked by being soaked in 1.5% BSA (0.5 - 1 h, RT) and were washed by PBS-Tween20 (2 minutes). In the next step, nitrocellulose membrane was incubated by primary antibody Anti Poly-Histidine-HRP (Sigma) (1:2000 diluted in BSA 1%) for 30 minutes at RT and it was washed again by PBS- Tween20 (3 × 5 minutes). Then it was incubated with a secondary antibody conjugated with HRP for 30 minutes at RT. After washing three times with PBS- Tween20 for 5 minutes, it was stained by Diaminobenzidine (0.05%DAB, 0.015% H2O2, 0.01MPBS, pH 7.2). Finally, the total concentration of our recombinant protein was assessed using Bradford process. In this regard, five dilutions of BSA protein, as the standard protein, were employed in 595 nm absorbance. The purified rCIL2-3E protein was saved at - 20°C for further immunogenicity study and its protective efficacy in mice model.
4. Results
4.1. In-Silico Conformation of Recombinant Construction
Reading framework of recombinant structure was confirmed using the CLC work bench software (Figure 1). In-silico study of the immunity profile of rCIL2-3E by VaxiJen server with a threshold of 0.5 had introduced this construct as an antigen with a score of 1.6328. Subsequently, the prediction of the second and third structure of the designed construction using I-tasser server showed that this structure included 42.5% Alpha helix structure and 33% random coil, 13% extended strand and 5.11% beta turn. Furthermore, the prediction of the third structure of the recombinant protein was done using I-tasser and PyMOL Vl software (Figure 2).
4.2. PCR Amplification, Cloning and Nucleotide Sequencing Analysis
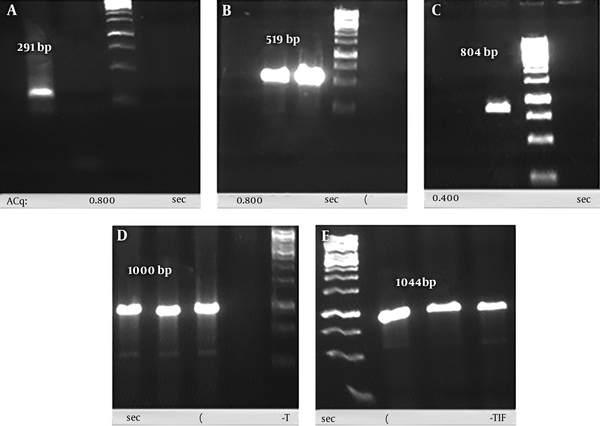
Gene codon of each subunit (291bp and 519bp) of the recombinant chimera sequences was amplified from synthesized sequences in PGH vector. The reproduction accuracy of each subunit was confirmed by agarose gel electrophoresis (Figure 3A and 3B). The SOE-PCR reaction with the purpose of connecting these two PCR products was performed successfully to produce rCIL2-3E (804 bp) (Figure 3C). Sub-cloning of this gene was conducted using the pTZ57R/T vector in DH5α strain of Escherichia coli as the host. Positive transformed colonies were confirmed using colony-PCR process and M13 universal primer, in which about 200 nucleotides were added to each sequence (Figure 3D). Ligation process of chimera fragment into pET-22b (+) vector as an expression vector, and its transformation to BL21 (DE3) strain of E. coli were successfully done. The integrity of the recombinant plasmids were confirmed using restriction enzyme double digestion (NcoI and XhoI) and also colony-PCR process using T7 universal primer (Figure 3E).
A, PCR products of 3 repeatation of OMP31 epitope; B, PCR products of IL-2; C, PCR products of rCIL2-3E using SOE-PCR procedure; D, conformation of inserting the rCIL2-3E into PTZ vector through colony-PCR process using M13 universal primer; E, conformation of inserting the rCIL2-3E into pET-22b (+) vector through colony-PCR process using T7 universal primer
4.3. Expression and Purification of Recombinant Protein
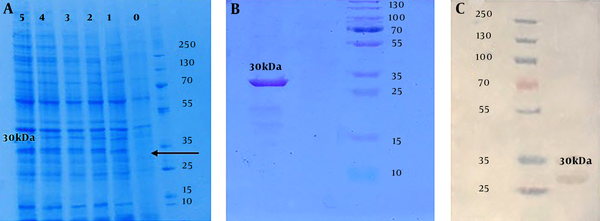
The expression of the target protein, induced in mid-log phase (OD600= 0.6) culture by adding 0.2 mM IPTG for 5 hours at 37°C, was detected through SDS-PAGE (12%) analysis (Figure 4A). The purification of produced target protein was successfully done using Ni-NTA affinity chromatography column. The accuracy and quality validation of the protein expressing approximately 30 kDa molecular mass, was approved using SDS-PAGE (12%) (Figure 4B). Western blotting assay with Anti Poly-Histidine-HRP antibody revealed specific reactivity with purified chimera protein produced in E. coli cells (Figure 4C). The estimated concentration of purified protein by Bradford assay (R² = 0.989) was 0.3 g/L.
Conformation of protein expression results: A, SDS-PAGE analysis of the rCIL2-3E protein with different sampling times after inducing by 0.2 mM IPTG; each column refers to yielded protein after 0, 1, 2, 3, 4 and 5 hours after induction by IPTG, respectively. B, SDS-PAGE analysis which approves purification of 30 kDa chimera protein band, correctly, using Ni-agarose column. C, Western blot conformation of 30 kDa chimera protein by Anti Poly-Histidine-HRP antibody.
5. Discussion
Brucellosis is a chronic bacteria zoonotic disease caused by Brucella genus bacteria. This widespread disease emerged from using pollutant livestock products; consequently, it can cause many economic losses and threatens human health (13). The most common pathogenic species in human is related to Brucella melitensis (14). The development of recombinant DNA techniques along with protein engineering science could develop fourth-generation vaccines, which only use immunogenic component of the pathogens. These recombinant vaccines, through their ability to stimulate appropriate responses and immunization, can be considered as good alternatives compared to traditional vaccines (15). Brucella is able to circumvent the host’s innate and adaptive immunity and as an intercellular pathogen, it can disrupt T cell energy and TCL through different pathways. Therefore, further immunity studies on Brucellosis disease are required to increase the knowledge on the provision of new therapeutic vaccines (16). Brucella belongs to intracellular pathogens. Host protection against Brucella is through cell-mediated immunity; the antigen present cells by engulfing the pathogen, process them into their antigens and represent them via their MHC II to T helper lymphocytes with CD4+ marker and MHC I to cytotoxic T lymphocytes with CD8+ marker (17). On the other hand, IL-2 is known as the main T cell growth factor and promotes T cell immunity (18). Although its expression has been reported by CD8+ T cells, dendritic cells and thymic cells, IL-2 is mainly produced by activating CD4+ T cells (19).
Prokaryote expression system is affordable, safe, and convenient and it is widely employed to produce recombinant proteins, which do not have post translation modification (20). Previously, the use of this expression system has shown the desired and practical results in relation to the expression of recombinant proteins, particularly proteins, which involved in immunological studies against Brucellosis disease (12, 21). Human IL-2 is folded by three Cys residues including Cys-58, Cys-105, and Cys-125, which needs oxidation environment to formation (22). Therefore, nascent proteins should be translocated to periplasm of the bacteria as oxidative compartment (23). E. coli uses Sec, TAT, and SPR pathways to translocate nascent proteins to periplasm based on signal peptides at N terminal of the protein. In general, Sec and SPR pathways can direct both folding and accumulation of nascent proteins in periplasm compartment, whereas TAT pathway conducts folding of nascent proteins in cytoplasm then accumulate them in periplasm (24). Thereby, to achieve a folded protein, our designed immunologic construct was inserted after PelB sequence of pET-22b (+) vector, which was applied to express the protein. In fact, PelB is known as one of the strongest signal peptides to direct nascent protein to periplasmic compartment via Sec pathway (25). It should not be forgotten that using PelB can facilitate protein extraction, because periplasm compartment is much clearer than cytoplasm, therefore, the protein of interest can be easily purified. In this study, we have attempted to introduce a recombinant protein, which contained IL2 and tandem repeat sequences of OMP31 antigen Th1 epitope of Brucella melitensis, as an efficient alternative for conventional vaccines.
5.1. Conclusion
This study was conducted to construct an immunological chimera structure consisting of interleukin-2 and three replicates of the experimental OMP31-epitope (IL2-3E) of the Brucella melitensis. For this purpose, this structure was designed and expressed in prokaryote expression system; therefore, its immunogenicity properties as a subunit vaccine in animal models can be later studied.
References
-
1.
Seleem MN, Boyle SM, Sriranganathan N. Brucellosis: a re-emerging zoonosis. Vet Microbiol. 2010;140(3-4):392-8. [PubMed ID: 19604656]. https://doi.org/10.1016/j.vetmic.2009.06.021.
-
2.
Cassataro J, Estein SM, Pasquevich KA, Velikovsky CA, de la Barrera S, Bowden R, et al. Vaccination with the recombinant Brucella outer membrane protein 31 or a derived 27-amino-acid synthetic peptide elicits a CD4+ T helper 1 response that protects against Brucella melitensis infection. Infect Immun. 2005;73(12):8079-88. [PubMed ID: 16299302]. [PubMed Central ID: PMC1307072]. https://doi.org/10.1128/IAI.73.12.8079-8088.2005.
-
3.
Berzofsky JA. Intrinsic and extrinsic factors in protein antigenic structure. Science. 1985;229(4717):932-40. [PubMed ID: 2410982].
-
4.
Vizcaino N, Cloeckaert A, Zygmunt MS, Dubray G. Cloning, nucleotide sequence, and expression of the Brucella melitensis omp31 gene coding for an immunogenic major outer membrane protein. Infect Immun. 1996;64(9):3744-51. [PubMed ID: 8751924]. [PubMed Central ID: PMC174288].
-
5.
Wales JR, Baird MA, Davies NM, Buchan GS. Fusing subunit antigens to interleukin-2 and encapsulating them in liposomes improves their antigenicity but not their protective efficacy. Vaccine. 2005;23(17-18):2339-41. [PubMed ID: 15755624]. https://doi.org/10.1016/j.vaccine.2005.01.040.
-
6.
Werle M, Bernkop-Schnurch A. Strategies to improve plasma half life time of peptide and protein drugs. Amino Acids. 2006;30(4):351-67. [PubMed ID: 16622600]. https://doi.org/10.1007/s00726-005-0289-3.
-
7.
Tovey MG, Lallemand C. Adjuvant activity of cytokines. In: Gwyn D, editor. Vaccine Adjuvants. Springer; 2010. p. 287-309.
-
8.
Chen X, Zaro JL, Shen WC. Fusion protein linkers: property, design and functionality. Adv Drug Deliv Rev. 2013;65(10):1357-69. [PubMed ID: 23026637]. [PubMed Central ID: PMC3726540]. https://doi.org/10.1016/j.addr.2012.09.039.
-
9.
Amet N, Lee HF, Shen WC. Insertion of the designed helical linker led to increased expression of tf-based fusion proteins. Pharm Res. 2009;26(3):523-8. [PubMed ID: 19002568]. [PubMed Central ID: PMC3121561]. https://doi.org/10.1007/s11095-008-9767-0.
-
10.
nazifi N, Tahmoorespur M, Sekhavati MH, Haghparast A. Engineering of tandem repeat construction of experimental epitopes of OMP31 antigen from Brucella melitensis linked to interleukin2 as an adjuvant to produce recombinant vaccine for sheep. Seventh congress of animal science of Iran. Iran. 2016.
-
11.
Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press; 1989.
-
12.
Yousefi S, Sekhavati MH, Abbassi Daloii T. Cloning and molecular characterization of Omp31 gene from Brucella melitensis Rev 1 strain. ARI. 2016;71.
-
13.
Corbel MJ. Brucellosis: an overview. Emerg Infect Dis. 1997;3(2):213-21. [PubMed ID: 9204307]. [PubMed Central ID: PMC2627605]. https://doi.org/10.3201/eid0302.970219.
-
14.
Ashford DA, di Pietra J, Lingappa J, Woods C, Noll H, Neville B, et al. Adverse events in humans associated with accidental exposure to the livestock brucellosis vaccine RB51. Vaccine. 2004;22(25-26):3435-9. [PubMed ID: 15308369]. https://doi.org/10.1016/j.vaccine.2004.02.041.
-
15.
Yousefi S, Abbassi-Daloii T, Sekhavati MH, Tahmoorespur M. Evaluation of immune responses induced by polymeric OMP25-BLS Brucella antigen. Microb Pathog. 2018;115:50-6. [PubMed ID: 29253594]. https://doi.org/10.1016/j.micpath.2017.12.045.
-
16.
Skendros P, Boura P. Immunity to brucellosis. Rev Sci Tech. 2013;32(1):137-47. [PubMed ID: 23837372].
-
17.
Pappas G, Akritidis N, Bosilkovski M, Tsianos E. Brucellosis. N Engl J Med. 2005;352(22):2325-36. [PubMed ID: 15930423]. https://doi.org/10.1056/NEJMra050570.
-
18.
Nelson BH. IL-2, regulatory T cells, and tolerance. J Immunol. 2004;172(7):3983-8.
-
19.
Yang-Snyder JA, Rothenberg EV. Spontaneous expression of interleukin-2 in vivo in specific tissues of young mice. Dev Immunol. 1998;5(4):223-45. [PubMed ID: 9814581]. [PubMed Central ID: PMC2275993].
-
20.
Pope B, Kent HM. High efficiency 5 min transformation of Escherichia coli. Nucleic Acids Res. 1996;24(3):536-7. [PubMed ID: 8602370]. [PubMed Central ID: PMC145660].
-
21.
Naghavi M, Sekhavati MH, Tahmoorespur M, Nassiry MR. Design and Production of a Novel Recombinant Chimeric IL2-Omp31 Antigen Against Brucella Infection. IRI. 2018;Inpress. https://doi.org/10.22092/ARI.2017.110504.1131.
-
22.
Kreitman RJ. Immunotoxins in cancer therapy. Curr Opin Immunol. 1999;11(5):570-8.
-
23.
Rosano GL, Ceccarelli EA. Recombinant protein expression in Escherichia coli: advances and challenges. Front Microbiol. 2014;5:172. [PubMed ID: 24860555]. [PubMed Central ID: PMC4029002]. https://doi.org/10.3389/fmicb.2014.00172.
-
24.
de Marco A, Deuerling E, Mogk A, Tomoyasu T, Bukau B. Chaperone-based procedure to increase yields of soluble recombinant proteins produced in E. coli. BMC Biotechnol. 2007;7:32. [PubMed ID: 17565681]. [PubMed Central ID: PMC1904446]. https://doi.org/10.1186/1472-6750-7-32.
-
25.
Forouharmehr A, Nassiri M, Govvati S, Javadmanesh A. Evaluation of Different Signal Peptides for Secretory Production of Recombinant Bovine Pancreatic Ribonuclease A in Gram Negative Bacterial System: an In silico Study. Curr Proteomics. 2017;14.