Abstract
Background:
Neutropenia is a side effect of chemotherapy in children with cancer, among which, bacteremia is the most commonly documented infection. Microbiological pattern of organisms causing bacteremia differs from one medical center to another. Due to the lack of comprehensive studies in Iran on bacteremia and resistance pattern of involved bacteria in febrile neutropenic children with malignancy, this study intended to investigate bacterial factors, and their antibiotic susceptibility and resistance patterns in these children.Methods:
In a retrospective study in the Children’s hospital of Tabriz from October 2007 to October 2013, medical records of 96 children hospitalized for malignancy with febrile neutropenia and positive blood culture, were reviewed on account of isolated bacteria, and their antimicrobial susceptibility or resistance.Results:
The mean age of the patients was 4.96 ± 3.08 years, and 52.1% (50 children) of them were female. The majority of malignant cases were hematologic with a prevalence of 60.4%, consisting of 35.4% Acute Lymphoblastic Leukemia (ALL), 15.6% Acute Myeloid Leukemia (AML), and 10.4% neuroblastoma. Among the isolated organisms from blood culture, 67.7% and 32.3% were Gram-positive and Gram-negative bacteria, respectively. Coagulase-negative staphylococci (28.1%), Staphylococcus aureus (24%), and Klebsiella pneumoniae (9.4%) were the most common isolated organisms. The rate of antibiotic susceptibility for Gram-negative organisms to ampicillin, gentamicin, ceftriaxone, ceftazidime, amikacin, imipenem, and ciprofloxacin were 61.5%, 66.7%, 37.9%, 16%, 61.5%, 85.2%, and 93.3%, respectively. In addition, 88% of coagulase-negative staphylococci and 77.3% of S. aureus isolates were oxacillin resistant. All cases of S. pneumoniae and Viridans streptococcus, and 50% of non-group A, B or D streptococci were penicillin resistant.Conclusions:
The maximum bacterial resistance was observed against older antibiotics. High rate of antibiotic resistance in most organisms, especially coagulase-negative staphylococci and Staphylococcus aureus, is a serious warning to promote the proper use of new antibiotics in empiric and prophylactic therapies.Keywords
Cancer Febrile Neutropenia Bacteria Antimicrobial Susceptibility
1. Background
Neutropenia is a side effect of chemotherapy in children with cancer. Such patients are more prone to infection, due to their abnormal mucous membranes, various catheters, poor nutrition, long-term exposure to antibiotics, and frequent hospitalization (1). Despite significant decrease in infection-caused mortality over the past decade, infection continues to be a major cause of death and illness in patients with cancer (2, 3). Understanding the epidemiology and drug resistance patterns of organisms causing infection in these specific patients, can greatly help physicians in making early and more effective decisions on the selection of empiric, inpatient or outpatient treatment to reduce mortality and disease rates (2, 4-9). Both Gram-positive and Gram-negative organisms are commonly isolated from blood samples of children with febrile neutropenia (3, 10). Over the past decade, changes in microbiological spectrum from Gram-negative to Gram-positive bacteria were well documented (11). This microbiological pattern may differ in various health centers (3, 5). Usually, the most common isolated Gram-positive pathogens are Viridans streptococcus, coagulase-negative staphylococci and Staphylococcus aureus (including MRSA) (3, 10). Nevertheless, Gram-negative organisms continue to be the main cause of bacteremia in developing countries, and are associated with high mortality rate (5). Several factors such as the difference in chemotherapy and prophylaxis regimens, use of catheters in central veins, hospital environment, and climate of the region affect common pathogens and their antibiotic resistance pattern. Considering these factors will contribute to the selection of appropriate initial antibiotics (12). There are a few studies from Iran on the causes of bacteremia in cancer patients, especially children, with febrile neutropenia, and also on their microbiological and antimicrobial resistance patterns. For example, in both of the mentioned studies, Gram-negative bacteria had the dominant pattern, yet, their antibiotic resistance or susceptibility was not addressed (13, 14).
This study intended to provide a precise evaluation of bacterial etiologic factors and measure antibiotic resistance to febrile neutropenia in children with cancer. The underlying reasons for this study were the lack of a comprehensive study on bacteremia, its causes, and resistance pattern of the involved bacteria in Iranian children with neutropenia, and the difference between the involved bacteria and their antibiotic resistance patterns in various medical centers.
2. Methods
2.1. Study Population
In this descriptive-retrospective study, medical records of children with malignancy, hospitalized at the Children’s hospital of Tabriz (the main children’s referral center in northwestern Iran) for febrile neutropenia and positive blood culture, from October 2007 to October 2013, were elicited and reviewed. Eligible patients were children with documented fever (+38°C for at least 1 hour), documented neutropenia (an absolute neutrophil count of < 1500/mL), and positive blood culture in terms of bacterial organisms. In addition, demographic information such as age, gender, length of hospitalization, fever level, initial diagnosis of illness, and disease duration, as well as paraclinical findings such as white blood cells and neutrophil counts, Erythrocyte Sedimentation Rate (ESR), and C-Reactive Protein (CRP) were elicited and analyzed. Accurate information of the isolated bacteria from the patients’ blood culture along with their antibiotic sensitivity or resistance was gathered.
2.2. Blood Culture Results and Identification of Bacteria
Blood samples of patients were cultured and processed using the automated BACTEC 9120 system (BD Diagnostic). Subcultures were performed from positive bottles onto chocolate agar. The obtained isolates were further processed as per the standard procedures to identify the pathogens (15).
2.3. Antimicrobial Susceptibility Testing
The antibiotic susceptibility of bacterial isolates was determined by the disc diffusion method (Kirby-Bauer), according to the Clinical and Laboratory Standards Institute (CLSI) recommendations (16). Escherichia coli ATCC 25922 and Staphylococcus aureus ATCC 25923 were used as control strains. Muller-Hinton agar medium for antimicrobial testing and antibiotic discs were obtained from Hi-media Laboratories, Mumbai, India.
2.4. Statistical Analysis
All research findings were analyzed using the SPSS software, version 22. For statistical investigations, descriptive statistical methods (frequency, percentage, and mean ± standard deviation) were used. To compare quantitative findings, the chi-square test was employed. In addition, for between-group comparison of the quantitative findings, the independent t test was utilized. The P value < 0.05 was considered significant.
3. Results
In this study, 96 cases with positive blood culture for bacteria in febrile neutropenic patients with malignancy were studied. The mean age of the patients was 4.96 ± 3.08 years, with a median of 3.6 years. The youngest and oldest patients were aged two months and 13 years, respectively. Among them, there were 46 (47.9%) males and 50 (52.1%) females. Figure 1 presents different malignancies in the patients. According to the diagram, the majority of malignant cases were hematological with the prevalence of 60.4%, consisting of 35.4% Acute Lymphoblastic Leukemia (ALL), 15.6% Acute Myeloid Leukemia (AML), and 10.4% neuroblastoma. The mean duration of hospitalization was 25.52 ± 11.30 days, with a median of 26.00 days. The maximum and minimum hospitalization duration was 5 and 69 days, respectively.
Different Malignancies in the Patients
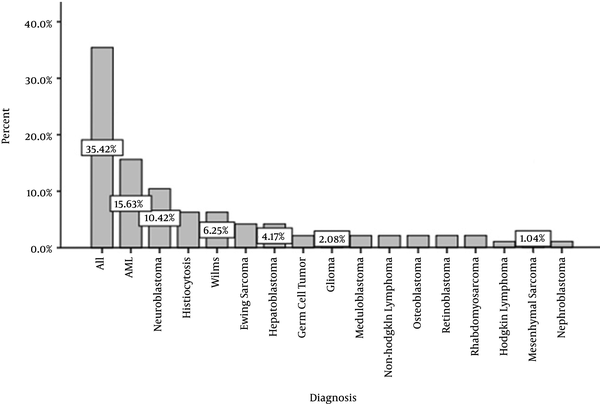
Fever and neutropenia were observed in 34 (35.4%) patients at the time of hospitalization. The mean duration of febrile neutropenia symptoms in the remaining patients was 9.47 ± 6.53 days, with a median of 7 days. The maximum and minimum duration of such symptoms was 1 and 28 das, respectively.
According to the laboratory findings, the majority of the patients had low White Blood Count (WBC) level and neutropenia. The hemoglobin and platelets were also at low levels; consequently, the majority of the patients had pancytopenia. In addition, 24% of patients were afflicted by profound neutropenia (absolute neutrophil count of < 100/mL). The mean level of ESR was 96.69 ± 61.68, with a median of 92. The maximum and minimum ESR levels were 15 and 305, respectively. The CRP levels were +1, +2, and +3 in 12 (12.5%), 31 (32.3%), and 11 (58.3%) cases, respectively.
Table 1 represents isolated microorganisms by their prevalence. Coagulase-negative staphylococci (28.1%), Staphylococcus aureus (24%), and Klebsiella pneumoniae (9.4%) were the most common isolated organisms with 27 (28.15%), 23 (24%), and 9 (9.4%) cases, respectively.
Frequency of Isolated Bacteria From Patient’s Blood Cultures
| Organisms | N. (%) |
|---|---|
| Coagulase-Negative Staphylococci | 27 (28.1) |
| Staphylococcus aureus | 23 (24) |
| Klebsiella pneumoniae | 9 (9.4) |
| Escherichia coli | 6 (6.3) |
| Non-fermentative Gram-negative bacilli | 6 (6.3) |
| Viridans Streptococci | 6 (6.3) |
| Acinetobacter spp. | 5 (5.2) |
| Non A, B, D Streptococci | 4 (4.1) |
| Enterococcus spp. | 3 (3.1) |
| Pseudomonas aeruginosa | 3 (3.1) |
| Streptococcus pneumoniae | 2 (2.1) |
| Capnocytophaga spp. | 1 (1) |
| Salmonella spp. | 1 (1) |
| Total | 96 (100) |
Among the isolated organisms, 67.7% and 3.32% were Gram-positive and Gram-negative bacteria, respectively. Amongst the Gram-positive organisms, 25.5% were resistant to vancomycin, 60.3% to cephalexin, 77.4% to ceftriaxone, 72.9% to cefotaxime, 81.7% to ceftizoxime, 83.6% to oxacillin, 95.2% to penicillin, 63.6% to ampicillin, 32.7% to rifampin, 79.9% to erythromycin, 65.1% to gentamicin, 55.7% to amikacin, 71.2% to co-trimoxazole, 41% to chloramphenicol, and 46.5% to ciprofloxacin. Amongst the Gram-negative organisms, the rates of resistance were as follow: imipenem (14.8%), cefixime (75%), ceftazidime (84%), cephalexin (90.5%), ceftriaxone (62.1%), cefotaxime (66.6%), ceftizoxime (67.8%), rifampin (33.3%), gentamicin (40%), amikacin (38.4%), chloramphenicol (51.8%), co-trimoxazole (57.1%), and ciprofloxacin (9.7%). Tables 2 and 3 represent the antibiotic susceptibility and resistance patterns for the isolates of Gram-positive and Gram-negative bacteria, respectively.
Antimicrobial Resistance Pattern of Gram-Positive Organisms
| Antibiotics | Pattern | Coagulase Negative Staphylococci | Staphylococcus aureus | Viridans Streptococci | Non A, B, D Streptococci | Enterococcus spp. | Streptococcus pneumonia |
|---|---|---|---|---|---|---|---|
| Vancomycin | S | 80.8% | 84.2% | 40.0% | 66.7% | 50.0% | 50.0% |
| R | 11.5% | 10.5% | 20.0% | 33.3% | 0.0% | 50.0% | |
| I | 7.7% | 5.3% | 40.0% | 0.0% | 50.0% | 0.0% | |
| Cephalexin | S | 26.1% | 56.5% | 50.0% | 33.3% | 0.0% | 0.0% |
| R | 69.6% | 43.5% | 50.0% | 33.3% | 100% | 100% | |
| I | 4.3% | 0.0% | 0.0% | 33.3% | 0.0% | 0.0% | |
| Ceftriaxone | S | 18.5% | 31.8% | 20.0% | 33.3% | 0.0% | 0.0% |
| R | 77.8% | 63.6% | 20.0% | 33.3% | 100% | 100% | |
| I | 3.7% | 4.5% | 60.0% | 33.3% | 0.0% | 0.0% | |
| Cefotaxime | S | 25.0% | 34.8% | 33.3% | 0.0% | 0.0% | 0.0% |
| R | 75.0% | 56.5% | 33.3% | 100% | 100% | 100% | |
| I | 0.0% | 8.7% | 33.3% | 0.0% | 0.0% | 0.0% | |
| Ceftizoxime | S | 15.4% | 28.6% | 0.0% | 25.0% | 0.0% | 0.0% |
| R | 80.8% | 71.4% | 66.7% | 50.0% | 100% | 100% | |
| I | 3.8% | 0.0% | 33.3% | 25.0% | 0.0% | 0.0% | |
| Oxacillin | S | 11.5% | 22.7% | 40.0% | 0.0% | 0.0% | 0.0% |
| R | 88.5% | 77.3% | 60.0% | 100% | 100% | 100% | |
| Penicillin | S | 0.0% | 4.3% | 0.0% | 50.0% | 0.0% | 0.0% |
| R | 100% | 95.7% | 100% | 50.0% | 100% | 100% | |
| Rifampin | S | 66.7% | 78.3% | 50.0% | 0.0% | 0.0% | 0.0% |
| R | 29.6% | 17.4% | 50.0% | 100% | 100% | 100% | |
| I | 3/7% | 4.3% | 0.0% | 0.0% | 0.0% | 0.0% | |
| Erythromycin | S | 7.7% | 36/8% | 33/3% | 25/0% | 0/0% | 0/0% |
| R | 80.8% | 57.9% | 66.7% | 75.0% | 100% | 50.0% | |
| I | 11.5% | 5.3% | 0.0% | 0.0% | 0.0% | 50.0% | |
| Clindamycin | S | 15.4% | 45.0% | 100% | 33.3% | 0.0% | 0.0% |
| R | 76.9% | 55.0% | 0.0% | 33.3% | 100% | 100% | |
| I | 7.7% | 0.0% | 0.0% | 33.3% | 0.0% | 0.0% | |
| Chloramphenicol | S | 53.8% | 66.7% | 83.3% | 33.3% | 33.3% | 50.0% |
| R | 30.8% | 23.8% | 16.7% | 33.3% | 33.3% | 50.0% | |
| I | 15.4% | 9.5% | 0.0% | 33.3% | 33.3% | 0.0% | |
| Co-trimoxazole | S | 20.0% | 27.3% | 33.3% | 0.0% | 66.7% | 0.0% |
| R | 80.0% | 63.6% | 66.7% | 66.7% | 33.3% | 100% | |
| I | 0.0% | 9.1% | 0.0% | 33.3% | 0.0% | 0.0% | |
| Ciprofloxacin | S | 60.0% | 57.1% | 50.0% | 33.3% | 33.3% | 0.0% |
| R | 36.0% | 33.3% | 25.0% | 66.7% | 33.3% | 100% | |
| I | 4.0% | 9.5% | 25.0% | 0.0% | 33.3% | 0.0% | |
| Ampicillin | S | - | - | 0.0% | - | 0.0% | - |
| R | - | - | 100% | - | 100% | - | |
| Amikacin | S | 61.5% | 47.6% | 16.7% | 0.0% | 0.0% | 0.0% |
| R | 26.9% | 42.9% | 83.3% | 100% | 66.7% | 100% | |
| I | 11.5% | 9.5% | 0.0% | 0.0% | 33.3% | 0.0% | |
| Gentamicin | S | 23.1% | 59.1% | 33.3% | 0.0% | 33.3% | 0.0% |
| R | 69.2% | 40.9% | 66.7% | 100% | 33.3% | 100% | |
| I | 7.7% | 0.0% | 0.0% | 0.0% | 33.3% | 0.0% |
Antimicrobial Resistance Pattern of Gram-Negative Organisms
| Antibiotics | Pattern | Salmonella spp. | Capnocytophaga spp. | P. aeruginosa | Acinetobacter spp. | NFGNB | E. coli | K. pneumoniae |
|---|---|---|---|---|---|---|---|---|
| Imipenem | S | 100% | 0.0% | 66.7% | 100% | 83.3% | 100% | 87.5% |
| R | 0.0% | 100% | 0.0% | 0.0% | 16.7% | 0.0% | 0.0% | |
| I | 0.0% | 0.0% | 33.3% | 0.0% | 0.0% | 0.0% | 12.5% | |
| Ceftazidime | S | 100% | 0.0% | 0.0% | 0.0% | 0.0% | 50.0% | 12.5% |
| R | 0.0% | 100% | 33.3% | 100% | 100% | 25.0% | 87.5% | |
| I | 0.0% | 0.0% | 66.7% | 0.0% | 0.0% | 25.0% | 0.0% | |
| Cefixime | S | 100% | 0.0% | 0.0% | 25.0% | 0.0% | 60.0% | 14.3% |
| R | 0.0% | 100% | 100% | 50.0% | 75.0% | 20.0% | 71.4% | |
| I | 0.0% | 0.0% | 0.0% | 25.0% | 25.0% | 20.0% | 14.3% | |
| Cefotaxime | S | - | 0.0% | 0.0% | 0.0% | 20.0% | 50.0% | 0.0% |
| R | - | 100% | 100% | 100% | 80.0% | 50.0% | 100% | |
| I | 100% | 0.0% | 0.0% | 40.0% | 33.3% | 75.0% | 33.3% | |
| Ceftriaxone | S | 0.0% | 100% | 66.7% | 40.0% | 50.0% | 0.0% | 66.7% |
| R | 0.0% | 0.0% | 33.3% | 20.0% | 16.7% | 25.0% | 0.0% | |
| I | 100% | 0.0% | 0.0% | 40.0% | 33.3% | 50.0% | 33.3% | |
| Cephalexin | S | 0.0% | 100% | 66.7% | 50.0% | 50.0% | 33.3% | 66.7% |
| R | 0.0% | 0.0% | 33.3% | 25.0% | 16.7% | 16.7% | 0.0% | |
| Ceftizoxime | S | 100% | 0.0% | 0.0% | 0.0% | 33.3% | 60.0% | 33.3% |
| R | 0.0% | 100% | 66.7% | 100% | 50.0% | 20.0% | 66.7% | |
| I | 0.0% | 0.0% | 33.3% | 0.0% | 16.7% | 20.0% | 0.0% | |
| Rifampin | S | - | 100% | - | - | - | - | - |
| R | - | 0.0% | - | - | - | - | - | |
| Chloramphenicol | S | 100% | 100% | 0.0% | 50.0% | 50.0% | 25.0% | 62.5% |
| R | 0.0% | 0.0% | 100% | 50.0% | 50.0% | 75.0% | 25.0% | |
| I | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 12.5% | |
| Co-trimoxazole | S | 100% | 0.0% | 0.0% | 25.0% | 83.3% | 50.0% | 33.3% |
| R | 0.0% | 100% | 100% | 75.0% | 16.7% | 50.0% | 66.7% | |
| Ciprofloxacin | S | 100% | 100% | 100% | 100% | 83.3% | 83.3% | 88.9% |
| R | 0.0% | 0.0% | 0.0% | 0.0% | 16.7% | 16.7% | 11.1% | |
| Amikacin | S | 100% | 0.0% | 100% | 100% | 33.3% | 75.0% | 50.0% |
| R | 0.0% | 100% | 0.0% | 0.0% | 50.0% | 25.0% | 50.0% | |
| I | 0.0% | 0.0% | 0.0% | 0.0% | 16.7% | 0.0% | 0.0% | |
| Gentamicin | S | 100% | 0.0% | 66.7% | 75.0% | 66.7% | 66.7% | 75.0% |
| R | 0.0% | 100% | 0.0% | 25.0% | 33.3% | 16.7% | 12.5% | |
| I | 0.0% | 0.0% | 33.3% | 0.0% | 0.0% | 16.7% | 12.5% | |
| Tobramycin | S | - | - | 66.7% | - | - | - | - |
| R | - | - | 0.0% | - | - | - | - | |
| I | - | - | 33.3% | - | - | - | - | |
| Carbenicillin | S | - | - | 33.3% | - | - | - | - |
| R | - | - | 33.3% | - | - | - | - | |
| I | - | - | 33.3% | - | - | - | - |
There was no statistically significant correlation between the isolated organism and gender (P = 0.62), age (P = 0.73), type of malignancy (P = 0.34), hospitalization duration (P = 0.1), neutrophil count (P = 0.94), lymphocyte count (P = 0.58), hemoglobin level (P = 0.06), platelets count (P = 0.83), ESR (P = 0.15), and CRP (P = 0.84).
4. Discussion
As mentioned earlier, bacteremia is a major cause of mortality among febrile neutropenic patients with cancer. In this study, 96 cases with positive blood culture for bacteria in febrile neutropenic children with malignancy were studies.
The mean age of the investigated patients was about 5 years, almost 1 year younger than cases presented in the studies of Hakim et al. (3), Castagnola et al. (10), and Gupta et al. (17), and 2 years younger compared to other studies (6, 10, 18, 19). This can be attributed to the methods used for identification and referral of the patients. Also, 47.9% of the patients were male, similar to the study of Al Omar et al. in Jordan (18). The gender difference was higher in similar studies conducted in European and American countries (3, 6, 10, 18, 19), which may be due to the regional epidemiological differences.
In addition, only 24% of patients were diagnosed with profound neutropenia (ANC >100 cells/mm3). This rate was 56% in the study of Hakim et al. (3), 82% in Gupta et al. (17), 60% in Paganini et al. (20), 76% in Sigurdardotti et al. (9), and 54% in Al Omar et al. (18), which can be attributed to differences in the employed methods and laboratory equipment.
In our study, about 60% of patients had hematological malignancies, which is almost similar to the findings of Hakim et al. (3) and Greenberg et al. (6), yet, lower (68%) than Paganini et al. (20), Gupta et al. (86%) (17), Sigurdardottir et al. (99%) (9), Tezcan et al. (64%) (19), and Al Omar et al. (66.9%) (18). This also can be partly due to epidemiological differences and systematic error in the sample selection.
In this study, 67.7% and 32.3% of the isolated bacteria from the patients’ blood cultures were Gram-positive and Gram-negative, respectively. Coagulase-negative staphylococci (28.1%), Staphylococcus aureus (24%), and Klebsiella pneumoniae (9.4%) were the most common isolated organisms with 27, 23, and 9 cases, respectively. In the study of Gupta et al., 61% of the isolated organisms were Gram-positive, in which coagulase-negative staphylococci accounted for the highest prevalence; whereas, Pseudomonas spp. were the most common Gram-negative organisms (17). In the study of Castagnola et al., in which 57% of the isolated organisms from blood culture were Gram-positive bacteria, coagulase-negative staphylococci strains were the most frequent isolates, followed by S. aureus. Among Gram-negative organisms, Escherichia coli and Klebsiella pneumoniae were the most frequently isolated bacteria (10). On the other hand, Al Omar et al. reported 75% Gram-positive bacteria with coagulase-negative staphylococci being dominant, and 25% Gram-negative organisms with Escherichia coli and Klebsiella pneumoniae being dominant (18). In the study of Hakim et al., 61% of cases were Gram-positive bacteria with Viridans streptococcus and coagulase-negative staphylococci being dominant, and 37% were Gram-negative organisms with Escherichia coli and Pseudomonas spp. being dominant (3). In contrast, Greenberg et al. reported 65% isolation rate for Gram-negative pathogens and only 30% Gram-negative pathogens, in which Klebsiella pneumoniae, coagulase-negative staphylococci, Pseudomonas spp., and Streptococcus spp. were the most common organisms (6). Moreover, Ariffin et al. (2000) reported that 61.9% of the isolated bacteria were Gram-negative organisms (21). This domination-difference of Gram-negative bacteria in the last two studies can be attributed to the time of studies (almost ten years ago) and current shift of etiologic factors of infection to Gram-positive organisms, which is due to newer prophylactic treatment patterns. Prevalence differences in various bacterial species can be attributed to epidemiological differences of medical centers and societies.
Greenberg et al. reported the susceptibility for Gram-negative organisms to ampicillin, gentamicin, ceftriaxone, ceftazidime, amikacin, imipenem, and ciprofloxacin as 18%, 73.5%, 56%, 83%, 89%, 86.5%, and 92.5%, respectively (6). In this study, these rates were 61.5%, 66.7%, 37.9%, 16%, 61.5%, 85.2%, and 93.3% for ampicillin, gentamicin, ceftriaxone, ceftazidime, amikacin, imipenem, and ciprofloxacin, respectively. Thus, results were relatively similar except for ampicillin, ceftazidime, and amikacin. Sigurdardottir et al. found that 57% of coagulase-negative staphylococci were oxacillin resistant (9). Moreover, no oxacillin resistant S. aureus isolate was observed, whereas, in this study, 88% of coagulase-negative staphylococci isolates and 77.3% of S. aureus strains were oxacillin resistant. Sigurdardotti reported only five cases (33%) of simultaneous gentamicin- and penicillin-resistant coagulase-negative staphylococci; however, this rate was 65% (17 cases) in our study. According to Sigurdardottir et al., none of the Streptococcal species were penicillin-resistant, and none of the Gram-negative bacteria were gentamicin, meropenem, ceftazidime or ciprofloxacin resistant; however, based on our findings, all Streptococcus pneumoniae and Viridans streptococcus isolates, and half of non-group A, B, or D streptococci were penicillin resistant. In addition, among the Gram-negative organisms, 40%, 14.8%, 84%, and 9.7% of the isolates were gentamicin, imipenem, ceftazidime, and ciprofloxacin resistant, respectively.
On the other hand, Ariffin et al. (2002) reported the resistance for Gram-negative bacteria to ceftazidime, amikacin, and imipenem as 26.3%, 21.2%, and 0.7%, respectively. In addition, the resistance for Gram-positive bacteria to methicillin was reported as 26.3%. Moreover, no resistance to vancomycin was observed (22). In this study, the resistance for Gram-negative bacteria to ceftazidime, amikacin, and imipenem was 84%, 38.4%, and 14.8%, respectively; in addition, the resistance for Gram-positive bacteria to methicillin and vancomycin was 83.6% and 25.5%, respectively. Ariffin et al. (2000) reported that only about 52%, 55%, 9.5%, and 13% of Klebsiella pneumoniae isolates were resistant to ceftazidime, amikacin, gentamicin, and ciprofloxacin, respectively; in addition, all Klebsiella pneumoniae isolates were susceptible to imipenem (21); whereas, the resistance for Klebsiella isolates to ceftazidime, amikacin, gentamicin, and ciprofloxacin was 87.5%, 50%, 25%, 11.1%, and 87.5%, respectively. Furthermore, the susceptibility to imipenem was reduced to 87.5%, and 12.5% of isolates only had intermediate resistance. Indeed, except for ciprofloxacin, the resistance to other antibiotics was significantly higher in our study. Differences observed in antibiotic resistance pattern of the isolates between this and previous studies can be attributed to differences in the method and type of antibiotic treatments, epidemiological differences, and prevalence of resistant strains in the region.
4.1. Conclusion
In general, the results showed that, similar to many other countries, etiological pattern of infection has shifted to Gram-positive organisms in the study site. Additionally, a high resistance level for the majority of organisms, especially coagulase-negative staphylococci and Staphylococcus aureus (the most common isolated organism), to older antibiotics such as penicillin and oxacillin, and also a high prevalence of antibiotic resistance for Klebsiella isolates (the most common Gram-negative organism) to ceftazidime were found. This emphasizes the need for adoption of preventive strategies such as continuous monitoring of antibiotic resistance pattern of common isolates and use of these findings in proper administration of available antibiotics for empiric and prophylactic therapies more than ever.
References
-
1.
Kliegman RSJ, Geme J, Behrman R. Nelson Textbook of Pediatrics. 19 th ed. 2011. p. 851-8.
-
2.
Auletta JJ, O'Riordan MA, Nieder ML. Infections in children with cancer: a continued need for the comprehensive physical examination. J Pediatr Hematol Oncol. 1999;21(6):501-8. [PubMed ID: 10598661].
-
3.
Hakim H, Flynn PM, Knapp KM, Srivastava DK, Gaur AH. Etiology and clinical course of febrile neutropenia in children with cancer. J Pediatr Hematol Oncol. 2009;31(9):623-9. [PubMed ID: 19644403]. https://doi.org/10.1097/MPH.0b013e3181b1edc6.
-
4.
Crokaert F. Febrile neutropenia in children. Int J Antimicrob Agents. 2000;16(2):173-6. [PubMed ID: 11053804].
-
5.
Feld R. Bloodstream infections in cancer patients with febrile neutropenia. Int J Antimicrob Agents. 2008;32 Suppl 1:S30-3. [PubMed ID: 18778919]. https://doi.org/10.1016/j.ijantimicag.2008.06.017.
-
6.
Greenberg D, Moser A, Yagupsky P, Peled N, Hofman Y, Kapelushnik J, et al. Microbiological spectrum and susceptibility patterns of pathogens causing bacteraemia in paediatric febrile neutropenic oncology patients: comparison between two consecutive time periods with use of different antibiotic treatment protocols. Int J Antimicrob Agents. 2005;25(6):469-73. [PubMed ID: 15890499]. https://doi.org/10.1016/j.ijantimicag.2005.01.020.
-
7.
Pizzo PA, Walsh TJ. Fungal infections in the pediatric cancer patient. Semin Oncol. 1990;17(3 Suppl 6):6-9. [PubMed ID: 2191445].
-
8.
Roguin A, Kasis I, Ben-Arush MW, Sharon R, Berant M. Fever and neutropenia in children with malignant disease. Pediatr Hematol Oncol. 1996;13(6):503-10. [PubMed ID: 8940733].
-
9.
Sigurdardottir K, Digranes A, Harthug S, Nesthus I, Tangen JM, Dybdahl B, et al. A multi-centre prospective study of febrile neutropenia in Norway: microbiological findings and antimicrobial susceptibility. Scand J Infect Dis. 2005;37(6-7):455-64. [PubMed ID: 16012006]. https://doi.org/10.1080/00365540510038497.
-
10.
Castagnola E, Fontana V, Caviglia I, Caruso S, Faraci M, Fioredda F, et al. A prospective study on the epidemiology of febrile episodes during chemotherapy-induced neutropenia in children with cancer or after hemopoietic stem cell transplantation. Clin Infect Dis. 2007;45(10):1296-304. [PubMed ID: 17968824]. https://doi.org/10.1086/522533.
-
11.
Hughes WT, Armstrong D, Bodey GP, Brown AE, Edwards JE, Feld R, et al. 1997 guidelines for the use of antimicrobial agents in neutropenic patients with unexplained fever. Infectious Diseases Society of America. Clin Infect Dis. 1997;25(3):551-73. [PubMed ID: 9314442].
-
12.
Mandell GL, Bennett JE, Dolin R. Mandell, Douglas, and Bennett's Principles and Practice of Infectious Diseases. 7th ed. Philadelphia: Churchill Livingstone/Elsevier; 2010. p. 4028-37.
-
13.
Alborzi A, Cheriki S, Karimi M, et al. Survey Incidence Rate of Bacterial Innfection In Febrile Neutropenic Patient (under 15) admitted in Shiraz Medical Hospital (1379-1380). Iran J Infect DisTrop Med. 2004;9(2):4-10.
-
14.
Hosseini SMJ, Ranjbar R, Ghasemi H, et al. A Study on the Prevalence and Etiology of Fever in Patients Hospitalized with Fever and Neutropenia in Baqyiatallah Hospital during 1995-2005. J Ilam Uni Med Sci. 2006;14(3):8-11.
-
15.
Mahon CR, Lehman DC, Manuselis G. Textbook of Diagnostic Microbiology. 3rd ed. Philadelphia: Saunders; 2007. p. 505-12.
-
16.
Clinical and Laboratory Standards Institute (CLSI). Performance standards for antimicrobial susceptibility testing; 19th informational supplement (M100-S19). Wayne, PA: CLSI; 2011.
-
17.
Gupta S, Bonilla M, Gamero M, Fuentes SL, Caniza M, Sung L. Microbiology and mortality of pediatric febrile neutropenia in El Salvador. J Pediatr Hematol Oncol. 2011;33(4):276-80. [PubMed ID: 21516023]. https://doi.org/10.1097/MPH.0b013e31820ff632.
-
18.
Al Omar S, Nazer L, Alkayed K. A prospective study of febrile neutropenia in pediatric cancer patients in Jordan. J Pediatr Hematol Oncol. 2013;35(8):614-7. [PubMed ID: 23823118]. https://doi.org/10.1097/MPH.0b013e31829f3480.
-
19.
Tezcan G, Kupesiz A, Ozturk F, Ogunc D, Gultekin M, Yesilipek A, et al. Episodes of fever and neutropenia in children with cancer in a tertiary care medical center in Turkey. Pediatr Hematol Oncol. 2006;23(3):217-29. [PubMed ID: 16517538]. https://doi.org/10.1080/08880010500506719.
-
20.
Paganini HR, Aguirre C, Puppa G, Garbini C, Ruiz Guinazu J, Ensinck G, et al. A prospective, multicentric scoring system to predict mortality in febrile neutropenic children with cancer. Cancer. 2007;109(12):2572-9. [PubMed ID: 17492687]. https://doi.org/10.1002/cncr.22704.
-
21.
Ariffin H, Navaratnam P, Mohamed M, et al. Ceftazidime-resistant Klebsiella pneumoniae bloodstream infection in children with febrile neutropenia. Int J Infect Dis. 2000;4(1):21-5.
-
22.
Ariffin H, Navaratnam P, Lin HP. Surveillance study of bacteraemic episodes in febrile neutropenic children. Int J Clin Pract. 2002;56(4):237-40. [PubMed ID: 12074201].