1. Background
Staphylococcus aureus (SA) is a major causative agent of nosocomial infections (1). Also, SA causes respiratory infections as well as skin problems and food poisoning. Due to increased resistance, SA is a growing public health challenge (2). Methicillin-resistant Staphylococcus aureus (MRSA) isolates represents a group of SA becoming resistant to several antibiotics such as erythromycin, levofloxacin, tetracycline, clindamycin, mupirocin, gentamicin, trimethoprim, and/or doxycycline, but it is usually sensitive to vancomycin (3). Most MRSA infections identify in those who have been in hospitals and other facilities, such as nursing homes and dialysis centers that are usually associated with invasive procedures or devices such as surgery, intravenous tubing, or artificial joints; hence it's also named as healthcare-associated MRSA (HA-MRSA). Nevertheless, MRSA also occurs in healthy people who haven’t been hospitalized, known as community-associated MRSA (CA-MRSA), often begins with painful skin rashes. Such infections can transmit by skin-to-skin contact. Hence, students, wrestlers, childcare professionals, and people living in crowded places are among those who are at increased risk of CA-MRSA (4). The resistance of SA to methicillin is because of the presence of the mecA gene (5). It’s estimated that the mortality rate of invasive MRSA in the United State is about 25% (6). For example, in the city of New York, its mortality rate during the period of 2002 - 2007 has been estimated as 30% (7). Because of increased antimicrobial resistance, Scientists are looking for new therapeutic interventions, with a particular emphasis on bacteriophages as a viable option in treating bacterial infections either in tandem with or as a substitution for antibiotics (7, 8). Methicillin has been used to treat SA infections, despite increasing resistance of many species to methicillin, and to find alternative therapeutic interventions; in the present study, methicillin was selected as the target antibiotic.
Bacteriophage, also named phage, is a group of viruses that infect and replicate inside the host bacteria and finally lyse it. To penetrate the host cell, the phage requires a specific receptor (9). Nevertheless, after disruption of the host cells, phages also disappear. Phages attack a wide range of cells (10). It’s a long time that the idea of phage therapy, using phages to treat infections caused by bacteria, is pursuing (11, 12). In some Eastern European countries, phages are using to treat bacterial infections (13, 14). For example, Jensen et al. (15) discovered a new lytic phage against MRSA and then purified it to eliminate human MRSA from hard fabrics and surfaces. The main reason for using phage cocktails is the extent of their activity, compared to using a single phage. Besides, they can affect different bacterial species and maintain their activity in different environmental conditions. Combining phages can also affect many bacterial strains that may be responsible for similar diseases. In general, they have a greater potential for treating infections than single phage isolates (16).
2. Objectives
Since multi-drug resistance has significantly increased in many SA isolates, the current study aimed to isolate phages that have a lytic effect on isolated methicillin-resistant S. aureus from clinical human samples.
3. Methods
3.1. Phage Isolation and Purification
A modified standard method was used to enrich and isolate bacteriophages. In brief, 50 mL of urban sewage in Isfahan, Iran, was centrifuged at 8000 g for 10 minutes at 4°C. Fifteen milliliter of the supernatant was added to 50 mL of SA and cultured overnight in nutrient broth, followed by 24 hours of incubation at 37°C to the multiplication of possible staphlophages. Residual bacterial cells and debris were removed by centrifugation (8000 g, 10 min, and 4°C) and filtration with a 0.22 µm Millipore filter. To confirm the presence of bacteriophage in filtrate, 100 µL of overnight-cultured SA was mixed with 10 mL of soft brain heart infusion (BHI) broth (Darmstadi, Germany). Then, the solution was covered on the surface of 1.5% solid BHI agar (Unipath LTD, England) until solidify for 30 minutes. Afterward, 10 µL of the filtrate was spotted on the surface and incubated at 37°C overnight. Phage lytic effects on bacteria were confirmed by the appearance of transparent zones (plaques) on the media (17).
3.2. Phage Characterization by Transmission Electron Microscopy
Phage morphology was determined by transmission electron microscopy (TEM) (Philips, Netherlands) of the purified phage particles. The surface of a carbon-coated copper grid was covered by staphylophages, and then negative staining with 2% (w/v) uranyl acetate was performed. The stained grids were dried with air and visualized at an accelerating voltage of 100 kw (17). Based on morphological characteristics, phages were identified in the family level of classification and nomenclature.
3.3. Phage Cocktail Preparation
For phage cocktail preparation, 200 µL of each isolated phage from different families was added to 100 µL of overnight-cultured SA isolates (17).
3.4. Sample collection for isolation of S. aureus
The samples were collected from blood, sputum, urine, ear, eyes, nose, abscess, pleural fluid, peritoneal fluid, broncho-alveolar lavage (BAL), trachea, throat, wound, secretions of patients, and the hospital environment during 2018 - 2019. SA was detected in the samples collected from two main teaching hospitals (AL-Zahra and Namazi) in Isfahan and Shiraz (Iran), respectively.
Samples were put in a nutrient broth medium (Scharlau Chemie, S.A.), and transferred to the laboratory. Then, samples were cultured on the surface of Muller Hinton agar and blood agar (Unipath LTD, England), followed by 24 h of incubation at 37°C. SA isolates were identified by colony morphology, Gram staining, and results of biochemical assays, including the production of DNase, coagulase, and catalase (17). The presence of SA was confirmed by the polymerase chain reaction (PCR) method.
3.5. Antimicrobial Susceptibility
The antibiotic susceptibility profile of S. aureus isolates against methicillin (5 µg), clindamycin (20 µg), erythromycin (15 µg), cefoxitin (5 µg), ciprofloxacin (5 µg), tetracycline (30 µg), and gentamycin (10 µg) (Padtan Teb, Iran) was evaluated by the Kirby-Bauer disk diffusion method according to clinical and laboratory standards institute (CLSI) guidelines. After 24 h of incubation at 37°C, the diameters of the zones of inhibition around the disc was compared to the interpretative criteria recommended in the CLSI guidelines. The bacterial suspension of each sample was made and compared with the 0.5 McFarland turbidity standard. The bacterial suspension was cultured on Mueller-Hinton agar (Unipath LTD, England) plates and then incubated at 35°C for 18 hours. Afterward, the zones of inhibition were measured according to the CLSI protocol (18).
3.6. DNA Extraction
To find SA and mecA gene, the bacterial genomic DNA extraction was performed using the boiling method. Few colonies of pure isolated bacteria were placed into a microtube containing 100 µL of double-distilled water and then were heated at 100°C for 10 minutes. The solution was centrifuged at 7000 g for 10 min at 4°C and the supernatant that containing bacterial genomic DNA was used for PCR assay (19).
3.7. PCR Test
The PCR assay targets the Staphylococcus genus-specific 16S rRNA gene with forwarding staph primer: 5’-AACTCTGTTATTAGGGAAGAACA-3’ and reverse staph primer: 5’-CCACCTTCCTCCGGTTTGTCACC-3’ was performed to confirm the isolates (20).
DNA was amplified by the PCR method in a total volume of 25 µL, containing 1.5 mM MgCl2, 30 mM dNTPs, 2 units of Taq DNA-polymerase, 1X PCR buffer (50 mM KCl, 10 mM Tris-HCl), 0.24 mM primer, and 50 ng DNA. The presence of the mecA gene was detected by PCR with previously designed primers include forward primer: 5’-GTAGAAATGACTGAACGTCCGATGA-3’ and reverse primer: 5’-CCAATTCCACATTGTTTCGGTCTAA-3’ (20).
Thermal cycling condition for PCR process was as follow 94°C for 3 minutes; 30 cycles at 94°C for 30 seconds, 55°C for 30 second, and 72°C for 45 seconds. The final extension was at 72°C for 5 minutes. PCR product (5 µL) was then analyzed by agarose gel electrophoresis method after staining with Safe Stain and visualized by UV (21).
3.8. Determination of Phage Cocktail Effects
To evaluate the phage cocktail effects against SA strains, the prepared mixture was tested using the double-layer agar method (DLAM). Plates were incubated at 37°C for 24 hours and observed for plaque formation (17).
3.9. Statistical Analysis
Data were analyzed by descriptive analysis and analyzed in SPSS version 19.
4. Results
4.1. Phage Isolation and Identification
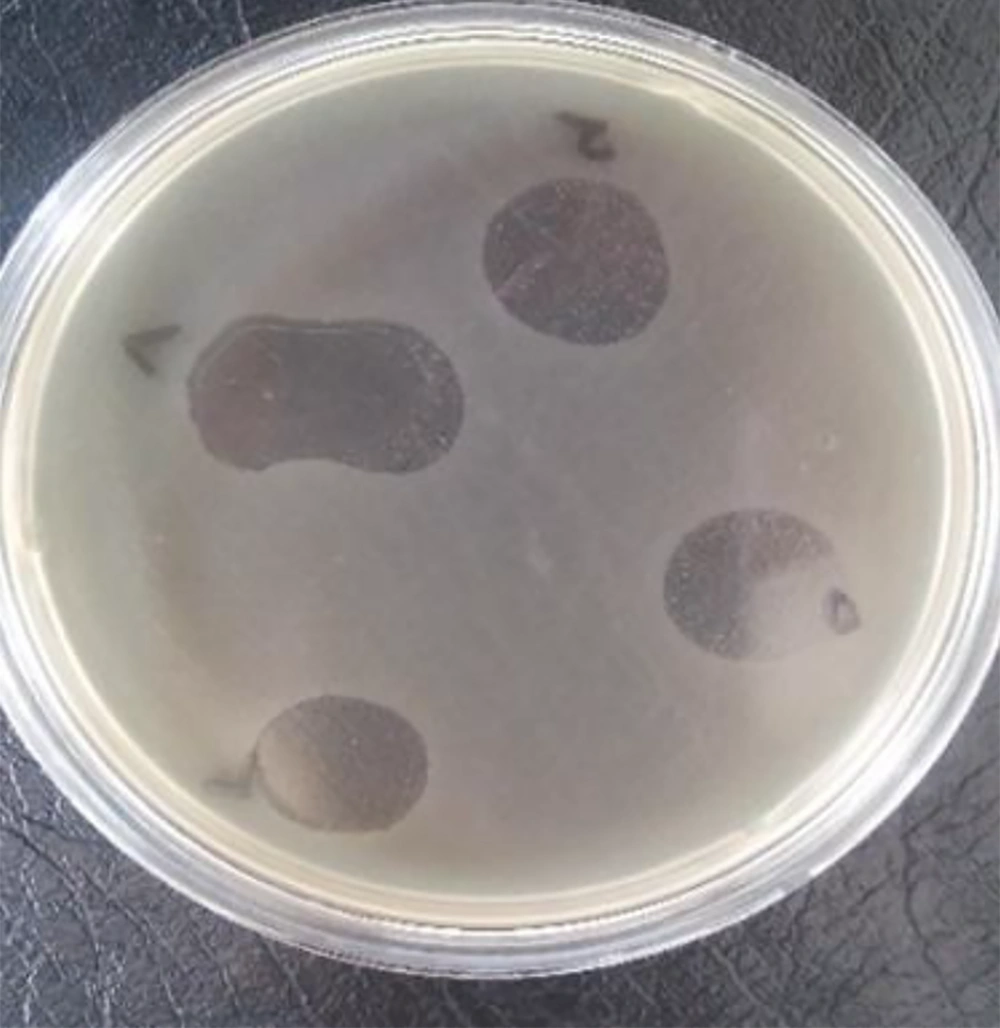
Lytic staphylophages were recognized by observing clear plaques on the double-layer plates (Figure 1).
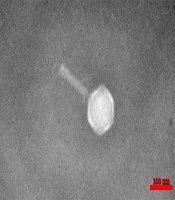
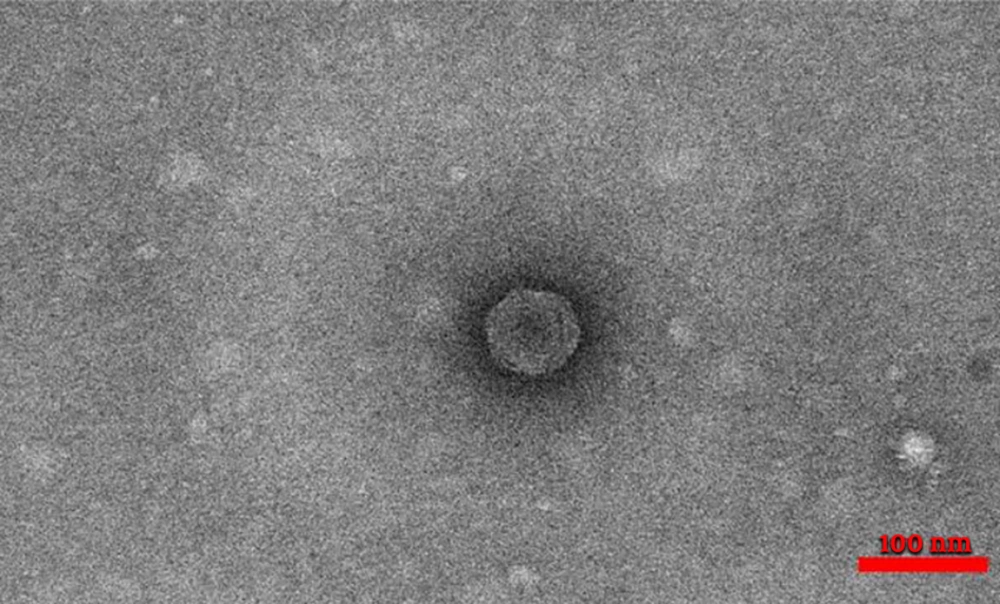
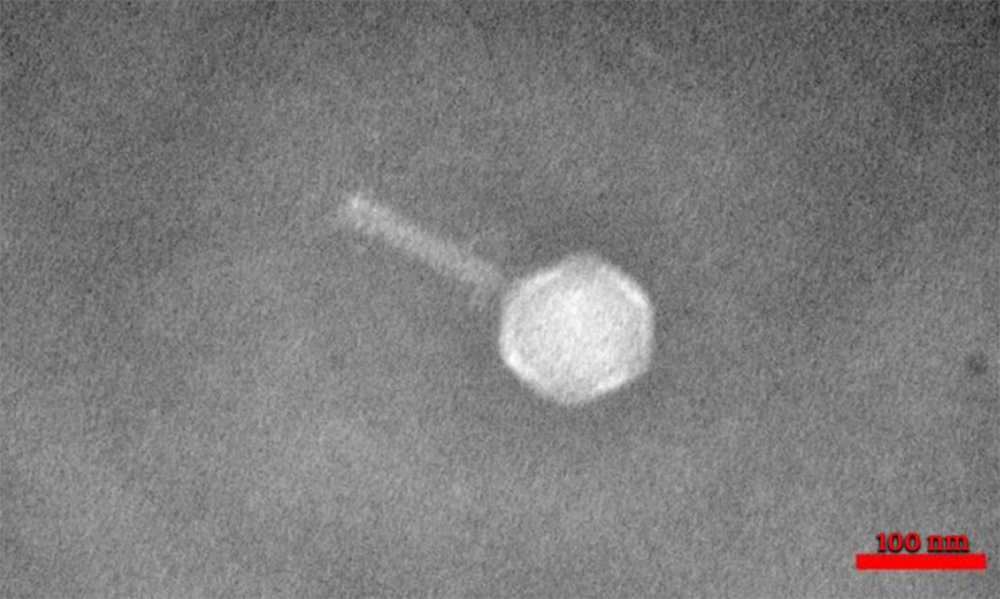
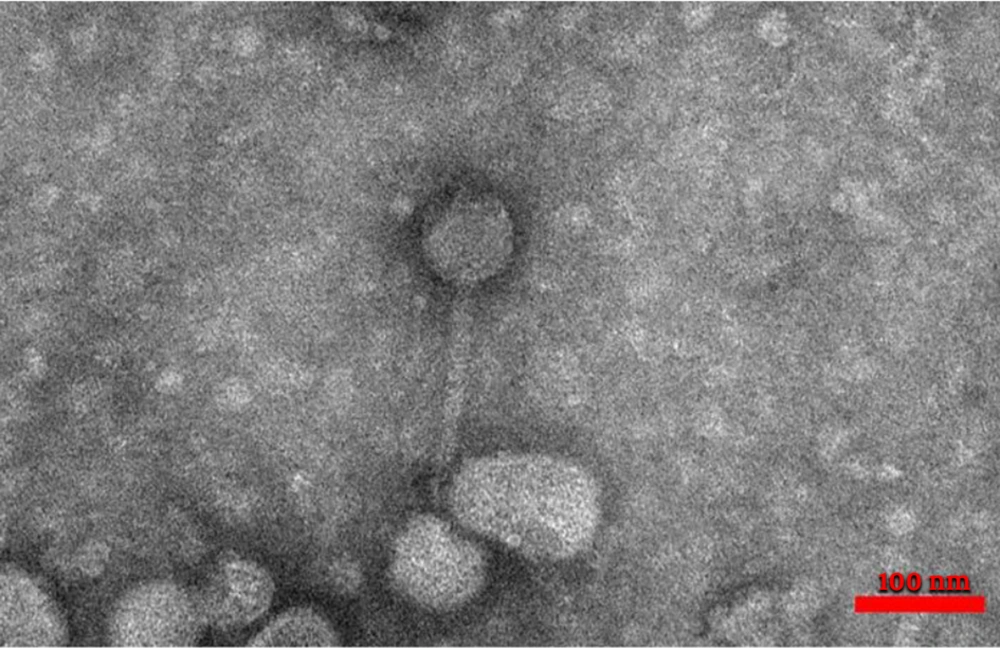
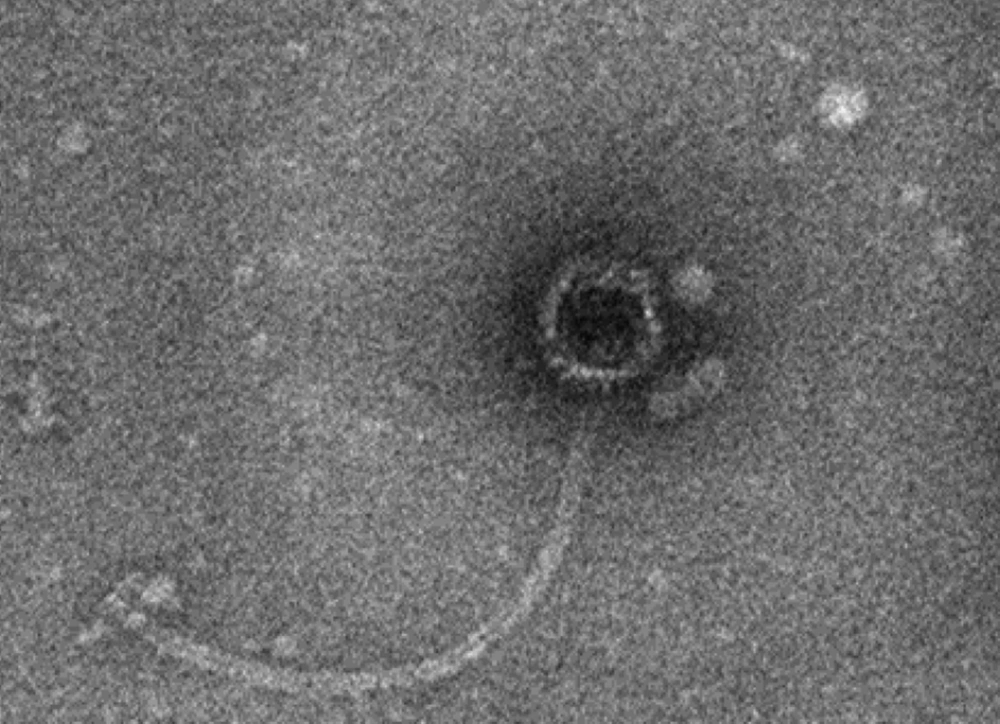
Four types of staphylophages were recognized by TEM and identified according to morphological characteristics (Springer index of viruses book) (22). The first type had a head with 30 nm width and 30 nm length, which was from the Tectiviridae family (Figure 2). The second phage had a head with 51 nm in width and 57 nm in length, a 60 nm tail, and a 15 nm endplate. This phage was from the Myoviridae family (Figure 3). For the other two phages, the width of the head was 30 and 33, and its length was 31 and 33 nm, while the tail was 78 and 160 nm tail, respectively. These phages were from the Siphoviridae family (Figures 4 and 5).
4.2. Antimicrobial Susceptibility Test and PCR
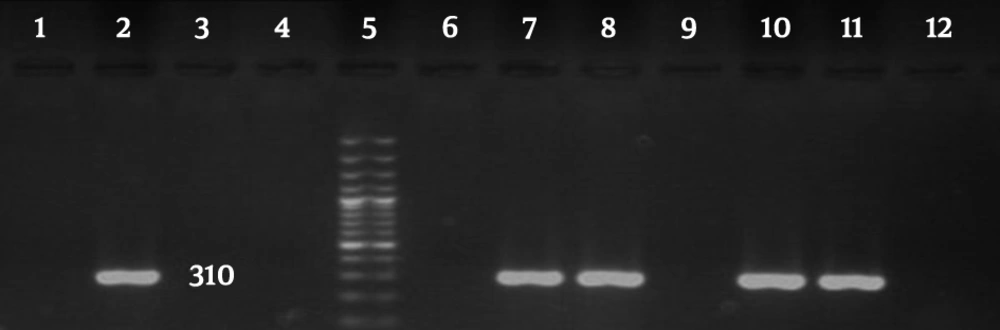
Based on the results of antibiotic susceptibility tests, 123 (92.4%) bacterial isolates (out of 133 Staphylococcus aureus isolates) were MRSA. Antimicrobial susceptibility profiles and the existence of the mecA gene are presented in Table 1. According to the PCR results, 118 (88.7%) samples were positive for the mecA gene (Figure 6).
| Antibiotic | Sensitive, No. (%) | Intermediate, No. (%) | Resistant, No. (%) | No. (%) |
|---|---|---|---|---|
| Methicillin | 3 (2.3) | 7 (5.3) | 123 (92.4) | |
| Clindamycin | 74 (55.6) | 15 (11.2) | 44 (33.0) | |
| Erythromycin | 74 (55.6) | 17 (12.7) | 42 (31.5) | |
| Cefoxitin | 50 (37.6) | 21 (15.7) | 62 (46.6) | |
| Ciprofloxacin | 50 (37.6) | 15 (11.2) | 68 (51.1) | |
| Tetracycline | 58 (43.6) | 17 (12.7) | 58 (43.6) | |
| Gentamycin | 91 (68.4) | 8 (6.0) | 34 (25.5) | |
| mecA gene | 118 (88.7) |
4.3. Staphylophage Cocktails and Phage Sensitivity Test
Two phage cocktails were prepared by mixing four types of staphylophage. Cocktail 1 contained phages VB-StaphT-Isf43 from the Tectiviridae family and VB-StaphS-Isf65 phage from the Siphoviridae family. Cocktail 2 contained VB-StaphM-Isf102 phage from the Myoviridae family and VB-StaphS-Isf 29 from the Siphoviridae family. The results of the phage susceptibility test showed that 44 SA isolates were lysed by two cocktails, and plaques were produced. Cocktails 1 and 2 lysed 19 (14.2%) and 25 (18.7%) MRSA isolates, respectively.
5. Discussion
In the present study, the resistance of clinical isolates of SA to methicillin was the highest reported rate in Iran, which indicates the growing trend during recent years. Faridi et al. (23) reported a low prevalence (10%) for MRSA among SA isolates. While Shokri et al. (2) reported a prevalence of about 57.8% in 45 SA isolates for methicillin resistance, and Rahimi et al. (24), 68% in 50 SA isolates. Safdari et al. (25) reported that 30% of 68 SA isolates were positive for beta-lactamase. Jafari-Sales et al. (26) reported that 75 (out of the 100 SA isolates) were methicillin-resistant, which 68% of them harbored the mecA gene. In the present study, the observed antibiotic resistance is the highest amount that is reported until now in Iran. Moreover, the prevalence of the mecA gene is the highest reported value in Iran. This difference can be attributed to factors such as geographical locations and the source of SA isolates. While phage therapy is a newly adopted approach in Iran, its application as a potential intervention to treat infections has a long history in some Eastern European countries. Phage susceptibility tests are used to identify the type of phages in some genus of bacteria, such as Brucella species. As mentioned above, most of the clinical isolates of S. aureus have become resistant to several drugs, including methicillin. Therefore, this study evaluated the antibacterial effect of staphylophages in eliminating the MRSA isolates as an alternative way to antibiotic therapy. Based on the findings, four types of staphylophage from Siphoviridae, Myoviridae, and Tecticoviridae families had a relatively good effect on MRSA clinical isolates. In this study, 44 samples (out of 133) were lysed by two phage cocktails, so that cocktails 1 and 2 lysed 19 (14.2%) and 25 samples (18.7%), respectively. Phage cocktails could lyse MRSA strains. Phage cocktail 2 (Myoviridae and Siphoviridae) had the highest lytic effect, which suggests its effectiveness. However, it needs further investigation in tissue cultures and animal models. Although some researchers reported that staphylophages could lyse MRSA isolates, but these effects may not be observed in isolates obtained from other geographical areas. In Eastern European countries and the former Soviet Union the potential antimicrobial effects of phage therapy are well-proved (27). Jensen et al. (15) isolated a new lytic phage against MRSA to eliminate infections caused by bacteria on the surfaces.
They realized that the fLizAnk phage had no toxic effect on fibroblast cell culture, and the antibacterial effect of phage against MRSA was presented in cell culture. Since the fLizAnk phage showed antibacterial activity against MRSA strains and had no cytotoxic effect against mammalian cells, it may be safe lonely or with a phage cocktail for the treatment of skin infection caused by SA. In another study by Jensen et al. (15) 12 phages were isolated and could replicate in human samples and/or MRSA isolates and eliminated the infection. They also investigated the advantages of some phages to decontaminate fomites (glass and cloth) and found a significant reduction in colony-forming units of MRSA following treatment with phage, including tests of a phage cocktail against MRSA isolates. In another study, Trigo et al. (28) used bacteriophages as an alternative antimicrobial treatment to control bacterial infections. Lehman et al. (29) have described the design and preclinical development of AB-SA01 (lytic myoviruses), a fixed-composition. Finally, they found that the inherent characteristics of AB-SA01 component phages met the regulatory and generally accepted criteria for human use, and reported that the presented preclinical data support the production under good manufacturing practices and phase 1 clinical studies with AB-SA01.
In another study, Abo-Elmaaty et al. (30) isolated a phage from the Myoviridae family and reported its antibacterial and anti-biofilm properties against SA. Jo et al. (31) designed a way to show the synergistic antimicrobial effect of phages combined with antibiotics against SA. Based on the results, the combined treatment of phages and antibiotics can be used to meliorate antimicrobial impression against antibiotic-resistant bacteria. Chan et al. (32) reported that phage therapy can be used to treat bacterial infections in humans, domestic animals, and even biocontrol in foods. Fabijan et al. (33), in a single-arm non-comparative trial on 13 patients with severe SA infections who were intravenously administered three Myoviridae bacteriophages (AB-SA01) as adjunctive therapy, reported no adverse reactions. This indicates that intravenous administration of AB-SA01 is safe in patients with severe SA infections, including infective endocarditis and septic shock.
5.1. Conclusions
This study demonstrated that some staphylophages isolated from urban sewage have appropriate effectiveness against clinical MRSA pathogens. Nevertheless, this issue should be further investigated in future studies on the inhibitory effects of staphylophages on other bacteria and even in vivo experiments.