Abstract
Background:
Dendrosomal nano-curcumin (DNC) represents an enormous potential to serve as a therapeutic anticancer agent. Here, we investigated the effect of DNC on wild type- and p53-mutant breast cancer cells.Methods:
MCF-7 and T47D cells were treated with DNC and investigated for cell viability through MTT assay. The mode of death was analyzed using Annexin V/FITC staining and PARP cleavage assays. Flow cytometric efflux and cell swelling tests were employed to assess the p-glycoprotein (P-gp) transporter activity. Moreover, real-time PCR was employed to study the expression of Bax, Bcl-2 as well as survivin and its ΔEx3 splicing variant.Results:
Our findings confirmed that DNC repressed cancer cell proliferation by induction of apoptosis. Additionally, DNC significantly modulated P-gp function that might lead to improved cellular permanence of curcumin. Malfunction of P-gp activity by DNC demonstrates its capability in reducing the drug resistance of p53-mutant cancer cells.Conclusions:
In conclusion, our findings demonstrated that dendrosomal nano-curcumin could be considered an anti-tumor therapeutic for p53-mutant tumor malignancies.Keywords
Curcumin Dendrosome Nanoparticles Apoptosis P-glycoprotein p53-mutant
1. Background
Despite recent advances in the treatment of cancers, tumor malignancy remains the second leading cause of death worldwide (1). Therefore, more effective therapeutic strategies are required. In recent years, scientists have re-focused on traditional medicine as a rich source for phytochemicals with potential anticancer properties (2). Curcumin, polyphenolic compound extracted from the rhizome of the plant Curcuma longa, has been employed as an antioxidant and anti-inflammatory remedy in traditional medicine (3-5). The compound has also been attributed to significant anticancer properties on numerous cancer cells, e.g., melanoma (6), colon (7), pancreatic (8), prostate (9), ovarian, and breast (10) in vivo and in vitro. Furthermore, curcumin does not significantly affect normal cells (11). Although the underlying molecular mechanisms of toxic effects of curcumin on cancerous cells are not well-known, different targets have been proposed that are modulated (12). Curcumin modulates the expression of different genes and/or proteins that play a major role in cell viability pathways, including NF-κB, Wnt/ β-catenin, and apoptosis (13).
Furthermore, curcumin can increase cellular accumulation of therapeutic compounds via modulating p-glycoprotein (P-gp) activity (14). Numerous findings have shown that curcumin downregulates membrane P-gp in both drug-resistant and non-resistant cancer cells that results in high cellular sensitivity and permeability of any therapeutic compounds, respectively (14, 15).
Nevertheless, the hydrophobic nature of this phytochemical strictly limits the bioavailability and cellular uptake (16). To overcome this obstacle, a promising approach is the encapsulation of curcumin in dendrosome nanoparticles that was recently formulated as dendrosomal nano-curcumin (DNC) in our lab (11, 17).
Based on our previous reports, dendrosomes are non-toxic, biodegradable, and spheroidal nanoparticles that could encapsulate curcumin and increase its stability (11). Compared to pristine curcumin, the physico-chemical and biological studies demonstrate the great potential of DNC to be a promising anticancer phytochemicals (18). Meanwhile, it has been proven that DNC could modulate cancer cell viability via induction of apoptosis (19). Here, we used MCF-7, with wild type p53 and T47D, harboring mutant p53, breast cancer cell lines to investigate the apoptotic potential of DNC and to elucidate whether a mutated p53 gene could influence cell death. Moreover, the effect of DNC on p-gp function was studied to evaluate whether this nano-compound modulates the function of P-gp and supports curcumin to be more stable inside the cells.
2. Methods
2.1. Reagents and Cell Lines
Human breast cancer cell lines, MCF-7 and T47D, were taken from Iranian national cell bank (Pasteur Institute, Tehran, Iran). The cells were grown in RPMI-1640 medium supplemented with 10% FBS, 5% CO2 atmosphere at 37°C, and 1% penicillin/streptomycin (all from Gibco, USA). Curcumin was purchased from Sigma-Aldrich, USA, and dendrosome nanoparticles were brought from Institute for Color Science & Technology, Tehran, Iran. The nano-based compound of curcumin; DNC was developed using an optimized protocol in our laboratory (11). The characteristics of DNC, including particle size, superficial charge (ζ-potentials) and encapsulation efficiency, were previously reported by Tahmasebi et al. (16). This work was approved by the ethics committee of the University of Tabriz as master thesis and submitted to IRANDOC with number 2292258.
2.2. Cellular Viability Assay
The viability of cells was measured by MTT assay on DNC-treated cells in a time- and dose-dependent manner. In brief, MCF-7 and T47D cells were seeded onto different 96-well plates in triplicate. After 12 hours, a serial concentration (0 - 120 µM) of pristine curcumin and DNC were added to the plates for 24, 48, and 72 h. Then, 20 µL MTT solution (5 mg/mL, Sigma-Aldrich, USA) was added to each well and incubated for three hours at 37°C. Reduced MTT was dissolved in 100 µl Dimethyl sulfoxide (DMSO) and measured spectrophotometrically at 570 nm by an Infinite M 200 pro microplate reader (Tecan, Switzerland). The IC50 values were determined by the linear interpolation method (11).
2.3. P-Glycoprotein Activity Assays
2.3.1. Flow Cytometric Efflux Assay
To assess a possible effect of DNC on the P-gp function, a well-known active transporter that conveys cytosolic compounds out of the cells, Rh123 efflux assay was performed in different groups, including control, DNC- and Verapamil (as positive control)-treated cells for 24, 48 and 72 h in dark conditions. After treatment of T47D and MCF-7 with either DNC or verapamil, they were incubated in 10 ng/µL Rhodamine 123 (Rh-123, Sigma-Aldrich, United states) for 60 min at 37°C. Then, treated cells were washed with ice-cold PBS and resuspended in 1 mL of medium for 90 min at 37°C. The intensity of intracellular trapped Rh-123 was evaluated by flow cytometry (BD FACSCalibur, Becton Dickinson, USA).
2.3.2. Cell Swelling Assay
To further decipher the malfunction of P-gp driven by DNC, T47D and MCF-7 cells were exposed to isotonic and hypotonic media. The isotonic medium was prepared in HEPES buffered saline solution (HBS), including 1 mM MgCl2, 4.5 mM KCl, 1 mM CaCl2, 95 m MNaCl, 5 mM HEPES, and 105 mM mannitol (pH = 7.3), while the hypotonic medium was mannitol-free. A number of 3 × 105 cells were seeded on 6-well plates and incubated with 60 µM DNC for 24, 48, and 72 h in a dark place. Then, adherent cells were washed with PBS and subjected to iso- and hypo-tonic conditions. Finally, the diameter of ten cells was measured randomly at different intervals of 20 and 60 min. All experiments were repeated at least three times, independently.
2.4. Apoptotic Assays
2.4.1. Annexin V/FITC
To determine the mode of death, Annexin V/FITC staining was carried out using the Affymetrix EBioscience kit. In brief, cancerous cells (3 × 105 cells/well) were seeded on 6-well plates, and after 12 hours, 60 µM DNC were added for 24, 48, and 72 h in a dark place. Then, the cells were trypsinized, collected by centrifugation, and resuspended in Annexin V-FLOUS solution at room temperature for 20 min. After washing with Binding Buffer, PI solution was added immediately prior to the study by flow cytometry (Becton Dickinson, Germany).
2.4.2. PARP Cleavage Assay
To further confirm apoptosis induced by DNC, Poly ADP-ribose polymerase (PARP) cleavage was studied using western blotting. In brief, MCF-7 cells were treated with 15 and 25 µM DNC for 24 h and prepared for protein extraction. Equal amounts of extracted proteins were fractionated on 10% SDS-PAGE gel and transferred onto a polyvinylidene fluoride membrane. After washing and blocking with 10% skim milk at room temperature for one hour, the membrane was incubated with PARP monoclonal antibody (1:1000, Cell Signaling, USA) at 4°C overnight. Moreover, β-actin was also employed as internal control (Santa Cruz, USA). The membrane was washed and incubated with horseradish peroxidase-conjugated anti-goat antibody (1:1000, Santa Cruz, USA) at room temperature for one hour. Finally, the bands were detected by 0.5 mg/ml diaminobenzidine (DAB) (Sigma-Aldrich, Germany) and 0.1% H2O2 in PBS.
2.5. Gene Expression Studies
2.5.1. RNA Extraction, DNase I Treatment and cDNA Synthesis
Quantitative real-time PCR (qRT-PCR) was employed to study the expression of apoptotic genes, including Bax, Bcl-2, BIRC5, and its ΔEx3 variant. Also, β2-microglobin (β2m) was used as internal control. MCF-7 and T47D cells were treated with 10, 30, and 50 µM DNC for 24, 48, and 72 h. Total RNA was extracted using TRizol® reagent (Invitrogen, USA), and DNase I enzyme (Fermentase, Lithuania) was used to remove any DNA contamination. Also, cycleScriptTM RT PreMix reagent (Takara Bio, Japan) was also applied for cDNA synthesis.
2.5.2. PCR
Semi-quantitative PCR was used to evaluate the expression of survivin ΔEx3, and qRT-PCR was employed for other genes mentioned in the previous paragraph. PCR for survivin ΔEx3 was done according to the manufacturer's instructions (Takara, Japan). Quantitative Real-time PCR was performed using 5 µL Syber Green Master Mix (Takara Bio, Japan), 250 nM of appropriate primers, and 200 ng of the generated cDNAs in appropriate thermocycler (Rotor-Gene Q, Germany) conditions. All reactions were done in triplicate. ΔCt (Ct relative Gene – Ct β2-microtubulin (as the internal control)) and ΔΔCt (ΔCt Treated – ΔCt Untreated) formulas were used in normalizing Ct values for each gene and to calculate the relative expression levels, respectively. The formula of 2-ΔΔCt was employed to plot the expression of genes (20). Table 1 has summarized the sequence of primers designed by Gene Runner V. 4.0.3.
The Primers Used for the Expression of the Studied Genes
| Gene | Primer | Size |
|---|---|---|
| Survivin-wt (wild type) (Real-time PCR) | 103 bp | |
| Forward | 5'- CGACCCCATAGAGGAACATA-3' | |
| Reverse | 5'-CCAGTTTCAAAAATTCACCAAG-3' | |
| Survivin (Semi-quantitative PCR) | 266 bp (survivin-wt) 148 bp (∆Ex3) | |
| Forward | 5'-CTTCATCCACTGCCCCAC-3' | |
| Reverse | 5'-CTTTCTCCGCAGTTTCCTC-3' | |
| Bax | 187 bp | |
| Forward | 5'- GCAAACTGGTGCTCAAGG -3' | |
| Reverse | 5'- ACTCCCGCCACAAAGA -3' | |
| Bcl-2 | 189 bp | |
| Forward | 5'-CGCATCAGGAAGGCTAGAGT-3' | |
| Reverse | 5'-AGCTTCCAGACATTCGGAGA-3' | |
| β2m | ||
| Forward | 5'- CTACTCTCTCTTTCTGGCCTG-3' | |
| Reverse | 5'- GACAAGTCTGAATGCTCCAC-3' | 191 bp |
| GAPDH | 493 bp | |
| Forward | 5'-AAGGTCATCCATGACAACTTTG-3' | |
| Reverse | 5'- GTCCACCACCCTGTTGCTGTAG-3' |
2.6. Statistical Analysis
The data were analyzed using the One-way ANOVA test, and values of P < 0.05 were accepted as statically significant. Also, IC50 values were obtained with non-linear regression using GraphPad PRISM (GraphPad Prism, RRID: SCR_002798) Version 6.01. Also, ImageJ software was employed to quantify the intensity of survivin ΔEx3 bands on agarose gel. All values were presented as the mean ± standard deviation of three independent assays.
3. Results
3.1. Effect of DNC on the Viability of MCF-7 and T47D Cells
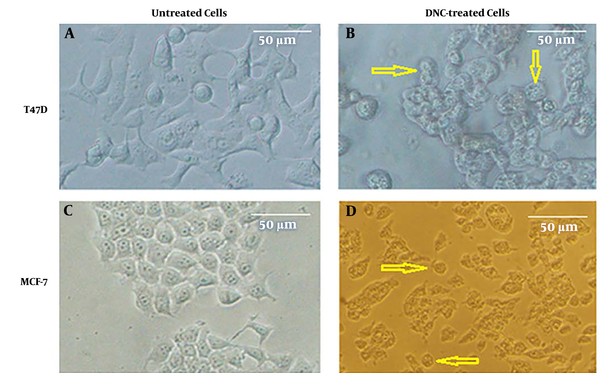
The cellular toxicity of DNC on breast cancer cell lines was studied using MTT assay. As Figure 1 shows, dendrosomal nano-curcumin significantly affected both cell lines morphologically. MTT assay demonstrated that DNC suppressed T47D and MCF-7 cells in a time- and dose-dependent manner (Figure 2 A and B). The IC50 for DNC in different time intervals of 24, 48, and 72 h for MCF-7 cells was obtained 83.89, 68.3, and 52.08 µM, respectively (Figure 2 C, P < 0.001). However, records for T47D cells were 116.2, 80.28, and 60.5 µM for 24, 48, and 72 h, respectively (Figure 2 B, P < 0.001).
Cytotoxic effect of DNC on MCF-7 and T47D cell lines; A and B show T47D cells before and after treatment with DNC, respectively. Similar data were taken for MCF-7 cells (C and D). Apoptotic cells are marked with arrows.
Cell viability of A: T47D and B: MCF-7 cell lines; C: The half-maximal inhibitory concentrations (IC50) of DNC at three different time intervals. Data are reported as Mean ± SD of three independent experiments. *: P < 0.05, **: P < 0.01.
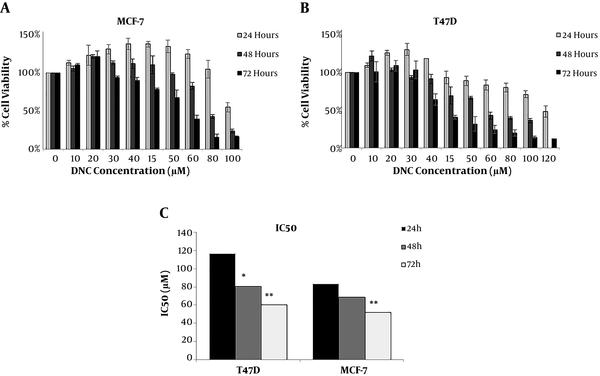
3.2. DNC Impairs P-Glycoprotein Function
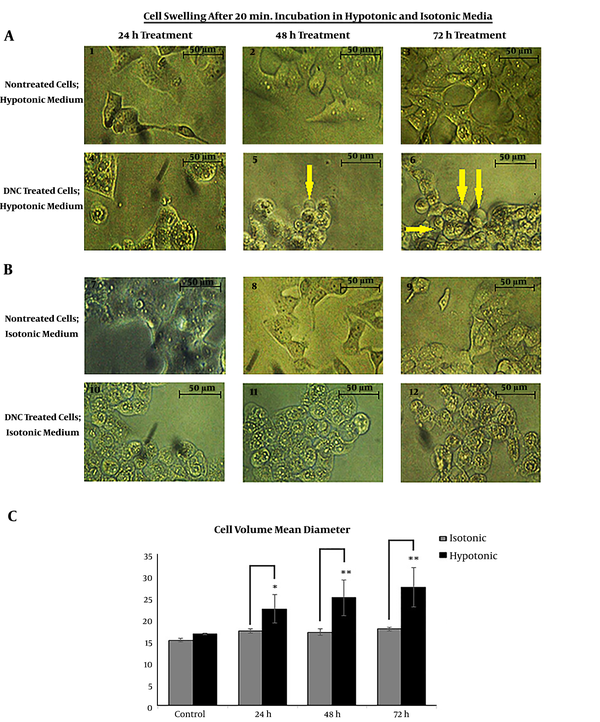
We assessed the DNC-caused malfunction of P-gp by cell swelling assay. To do this, both cells were treated with DNC in hypotonic and isotonic media. According to the microscopic records on Figure 3 A and B, the cell volume has obviously changed in DNC treated-T47D and MCF-7 cells. After 60 minutes, we calculated the cell diameter for T47D cells under both isotonic and hypotonic conditions (P Treatment time vs. cell diameter = 0.012 and P Cell diameter vs. medium condition = 0.001). It was found that changes in average cell volume driven by hypotonic medium were more significant than under isotonic conditions. Similar findings were recorded for MCF-7 (data not shown).
Cell swelling assay in hypotonic and isotonic media; A and B: Cell volume mean changes in T47D cells; Microscopic visualization confirmed changes in cellular volume as shown by arrows. C: The mean volume of DNC treated-T47D cells significantly increased under hypotonic condition (P Treatment time vs. cell diameter = 0.012 and P Cell diameter vs. medium condition = 0.001). The changes in cellular mean diameter in the isotonic condition is not significant (P = 0.15). Data are reported as mean ± SD of three independent experiments. *: P < 0.05, **: P < 0.01.
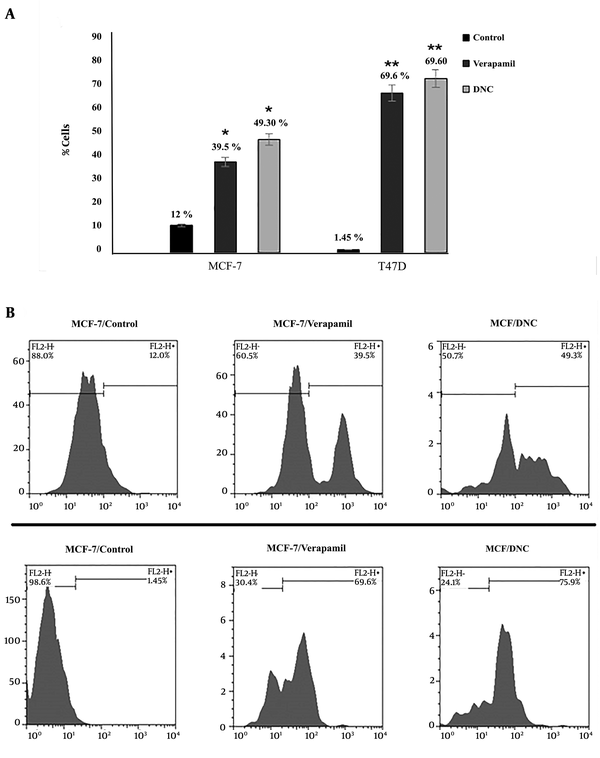
Efflux pump assay was also employed to detect the intensity of rodamine 123 MCF-7 and T47D cells over a period of 72 h (P Control versus DNC = 0.001; P Verapamil vs. DNC > 0.05). As Figure 4 shows, no significant effect was detected in verapamil, and DNC treated cells.
Efflux pump assay was used to evaluate the intensity of Rd 123 in DNC and verapamil treated cells. Flow cytometry detected a significant decrease in the efflux of Rh-123 in both DNC and Verapamil (as positive control) treated samples (P = 0.001). There were no significant differences between DNC and Verapamil function in deactivating P-gp. (P DNC vs. Verapamil > 0.05).
3.3. DNC Induces Apoptosis in Breast Cancer Cells
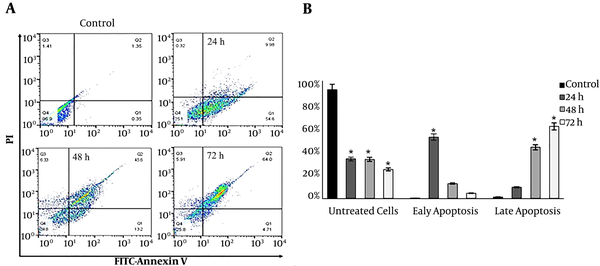
Annexin V/FITC and PARP cleavage assays were employed to study the mode of death in DNC treated cells. As Figure 5A illustrates, the treatment of T47D cells with 60 µM DNC induced apoptosis in a time-dependent manner (P < 0.05). The percentages of early apoptosis in treated cells were 54.6%, 13.2%, and 4.71% after 24, 48, and 72 h, respectively. In contrast, these findings for late apoptosis were 9.98%, 45.6%, and 64% for 24, 48, and 72 h, respectively (Figure 5B). Similar findings were detected for MCF-7 (data not shown).
Study of apoptosis by Annexin V/FITC; A: Annexin-V-FLOUS/PI staining in T47D cells after 24, 48, and 72 h treatment with 60 µM DNC. B: The percentage of cells in early and late apoptosis (P ≤ 0.01). Data are reported as Mean ± SD of three independent experiments. *: P < 0.05, **: P < 0.01.
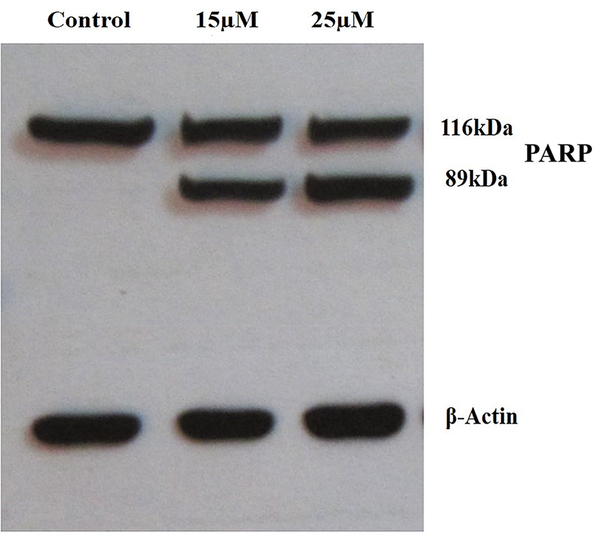
To further confirm the apoptotic effect of DNC, PARP cleavage assay was performed. The western blotting analysis showed that PARP protein is cleaved into two segments with 116 kDa and 89 kDa (Figure 6). As expected, fragmented PARP was not detected in untreated cells.
DNC induces apoptosis in MCF-7 cells treated with 15µM and 25µM for 48 h through caspase-dependent PARP cleavage. PARP protein is cleaved in DNC-treated cells.
3.4. Expression Profile of Pro- and Anti-Apoptotic Genes in DNC-Treated Cells
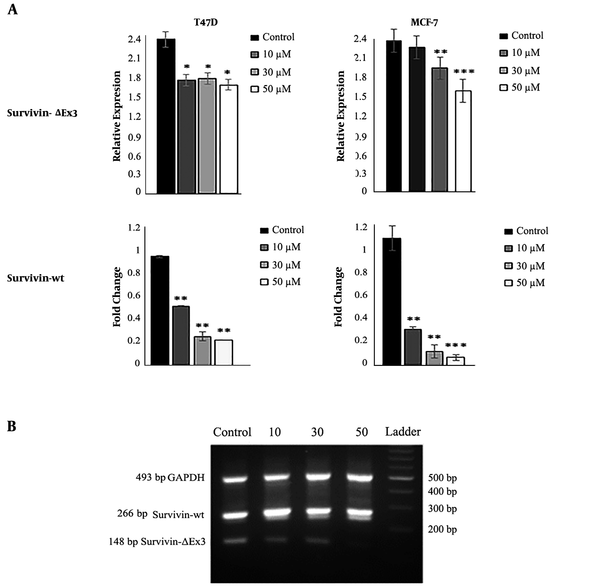
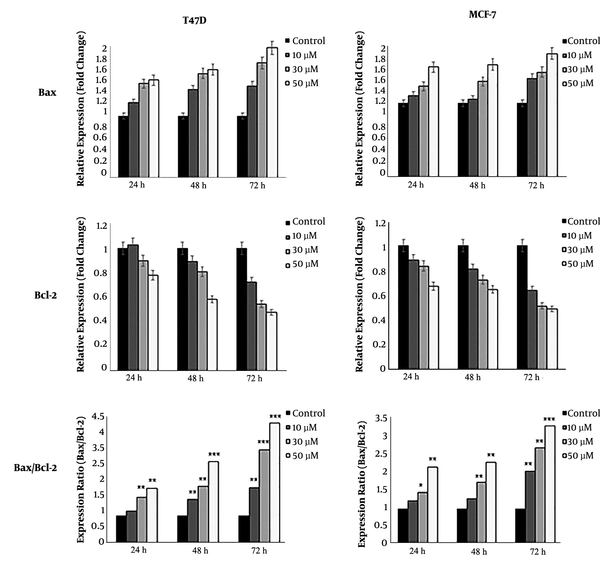
Real-time PCR detected the expression of all apoptotic and anti-apoptotic genes. Our results showed that DNC significantly decreased the expression of anti-apoptotic survivin and its spliced variant survivin-ΔEx3 in both MCF-7 and T47D cells (Figure 7). Agarose gel electrophoresis detected the expression of survivin and its variants in both cells lines that are shown in Figure 7 B for T47D. Furthermore, the expression ratio of Bax/Bcl2 as a hallmark of apoptosis was elevated in both cell lines (Figure 8).
The expression of survivin (real-time PCR) and its splicing variant survivin-ΔEx (semi-quantitative PCR) in MCF-7 and T47D cells. B: Visualization of survivin-ΔEx expression on 1.5% agarose gel electrophoresis. The intensity of bands was evaluated by ImageJ in comparison with internal control. Data represent the mean ± standard deviation of three independent experiments. * P < 0.05, ** P < 0.01 and *** P < 0.001.
A: The expression ratio of Bax/Bcl-2 in MCF-7 and T47D cells treated with DNC after 72 hours. B: Detection of the expression of survivin and its spliced variants on gel electrophoresis in TD7D cells. The intensity of bands was analyzed by ImageJ software to semi-quantify data. Note that only three variants have been marked on gel. Data represent the mean ± standard deviation of three independent experiments. *P < 0.05, **P < 0.01 and *** P < 0.001.
4. Discussion
Since current therapeutic agents for chemotherapy of many cancers are not completely effective, and still there are drastic challenges in this way, scientists make efforts on finding novel anticancer compounds to decrease these problems as well as possible (21). In recent decades, phytochemicals compounds attract much attention (22, 23). Curcumin is the active ingredient of turmeric that its pharmaceutical properties have been revealed in traditional medicine (24). Anticancer characteristic is the most important pharmaceutical property of this ingredient that has been approved in several studies in vitro and in vivo, but the molecular mechanism of its action is not completely clear yet (25, 26). On the other hand, the low stability and bioavailability of curcumin limit its clinical application (16). Therefore, several curcumin nano-structures have been developed that overcome these drawbacks (17, 27). Dendrosomal nano-curcumin (DNC) is one of those nano-structures that improve the solubility, cellular uptake, and toxicity of curcumin (11, 18). According to this study, we aimed to apply curcumin as a toxic agent for breast cancer cells. This is almost the first study to evaluate the ability of DNC in the modulation of P-gp function as a well-known active transporter that conveys cytosolic compounds out of the cells.
First, we report that DNC significantly affects the viability of MCF-7 and T47D cells in a time- and dose-dependent manner; whereas, pristine curcumin, as well as pristine Dendrosome, did not show any remarkable toxicity at the same concentrations as the nano-based compound (data for curcumin and dendrosome not shown). Therefore, only DNC was investigated in further assessments. However, we previously reported that DNC does not affect normal cells, including mouse fibroblasts and human peripheral blood mononuclear cells (11). Furthermore, we employed Annexin V/FITC assay and western blotting for PARP cleavage to detect the mode of death. As described in the results, DNC has induced apoptosis in T47D cells in a time-dependent manner. The effect of DNC on suppressing cancer cell proliferation via induction of apoptosis has been reported earlier for different cancerous cells (27-30).
Extensive studies have demonstrated that curcumin modulates the expression of numerous pro- and anti-apoptotic genes. It has also been shown that curcumin increases the expression of Bax and decreases the expression of Bcl-2 in both p53-mutant and p53 wild-type cells, which could confirm the ability of curcumin in promoting apoptosis through both p53 dependent and independent pathways (31). Our findings are in concordance with the aforementioned report that DNC significantly increased the expression ratio of Bax/Bcl-2 in treated cells (28).
In addition, we found that DNC downregulates the expression of anti-apoptotic survivin gene. This finding confirms Glienke et al. study who investigated the curcumin impact on the downregulation of survivin on pancreatic cancer cell lines (29). Survivin comprises various splicing variants, named survivin-wt, survivin-2α, survivin-2B, survivin-3B, and survivin-3ΔEx (31). Survivin-wt and survivin ΔEx3 are two well-known variants with anti-apoptotic function (32). Upregulation of these variants eliminates mitochondria-dependent apoptosis where Bcl-2/survivin-ΔEx3 complex inhibits caspase-3 activity (33). Curcumin has been shown to downregulate the expression of survivin in various types of cancer (34, 35). As our results illustrate, DNC significantly modulates the function of survivin and its variants that finally, apoptosis is induced in treated cells.
Furthermore, to partially uncover the reason for the high cellular accumulation of DNC versus pristine curcumin, efflux, and cell swelling assays were performed. Numerous works have shown that dysfunction of the P-gp membrane pump creates cell swelling, addressing its role in cell volume-regulated chloride conductance path (36). Our findings represent that fluorescent Rho-123 is significantly accumulated in DNC-treated breast cancerous cells as like as Verapamil (as positive control). Enhancement of intracellular Rh-123 is related to the capacity of DNC to modulate the function of P-gp trans-membrane pump. In addition, results from cell swelling assay illustrated that treating cells with DNC impairs cellular trans-membrane pumping protein P-gp. In fact, P-gp plays a major role in controlling cellular osmotic pressure when cells are exposed to the hypotonic medium. In many studies, resistance to the cytotoxic drugs has most often been linked to the overexpression of P-gp, and therefore, dysfunction of this membrane protein probably can reduce cellular resistance to chemotherapy (37-39).
4.1. Conclusion
This study not only demonstrates the apoptotic effect of DNC on p53-mutant T47D cells but also, for the first time, unravels the role of multidrug resistance P-glycoprotein in cellular permeability and stability of curcumin. Therefore, it may be inferred that our nano-based curcumin has the potential to be employed as therapeutic anti-tumor medicine even for p53-mutant tumors. However, further investigations on the effect of DNC on other death-related pathways and in vivo studies are required that some of them are in progress in our lab.
References
-
1.
Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65(1):5-29. [PubMed ID: 25559415]. https://doi.org/10.3322/caac.21254.
-
2.
D'Incalci M, Steward WP, Gescher AJ. Use of cancer chemopreventive phytochemicals as antineoplastic agents. Lancet Oncol. 2005;6(11):899-904. [PubMed ID: 16257798]. https://doi.org/10.1016/S1470-2045(05)70425-3.
-
3.
Yallapu MM, Jaggi M, Chauhan SC. Curcumin nanomedicine: a road to cancer therapeutics. Curr Pharm Des. 2013;19(11):1994-2010. [PubMed ID: 23116309]. [PubMed Central ID: PMC3640558]. https://doi.org/10.2174/138161213805289219.
-
4.
Menon VP, Sudheer AR. Antioxidant and anti-inflammatory properties of curcumin. Adv Exp Med Biol. 2007;595:105-25. [PubMed ID: 17569207]. https://doi.org/10.1007/978-0-387-46401-5_3.
-
5.
Mun SH, Joung DK, Kim YS, Kang OH, Kim SB, Seo YS, et al. Synergistic antibacterial effect of curcumin against methicillin-resistant Staphylococcus aureus. Phytomedicine. 2013;20(8-9):714-8. [PubMed ID: 23537748]. https://doi.org/10.1016/j.phymed.2013.02.006.
-
6.
Liao W, Xiang W, Wang FF, Wang R, Ding Y. Curcumin inhibited growth of human melanoma A375 cells via inciting oxidative stress. Biomed Pharmacother. 2017;95:1177-86. [PubMed ID: 28926928]. https://doi.org/10.1016/j.biopha.2017.09.026.
-
7.
Lee YH, Song NY, Suh J, Kim DH, Kim W, Ann J, et al. Curcumin suppresses oncogenicity of human colon cancer cells by covalently modifying the cysteine 67 residue of SIRT1. Cancer Lett. 2018;431:219-29. [PubMed ID: 29807115]. https://doi.org/10.1016/j.canlet.2018.05.036.
-
8.
Kesharwani P, Banerjee S, Padhye S, Sarkar FH, Iyer AK. Parenterally administrable nano-micelles of 3,4-difluorobenzylidene curcumin for treating pancreatic cancer. Colloids Surf B Biointerfaces. 2015;132:138-45. [PubMed ID: 26037703]. https://doi.org/10.1016/j.colsurfb.2015.05.007.
-
9.
Yang J, Wang C, Zhang Z, Chen X, Jia Y, Wang B, et al. Curcumin inhibits the survival and metastasis of prostate cancer cells via the Notch-1 signaling pathway. APMIS. 2017;125(2):134-40. [PubMed ID: 28120490]. https://doi.org/10.1111/apm.12650.
-
10.
Liu LD, Pang YX, Zhao XR, Li R, Jin CJ, Xue J, et al. Curcumin induces apoptotic cell death and protective autophagy by inhibiting AKT/mTOR/p70S6K pathway in human ovarian cancer cells. Arch Gynecol Obstet. 2019;299(6):1627-39. [PubMed ID: 31006841]. https://doi.org/10.1007/s00404-019-05058-3.
-
11.
Babaei E, Sadeghizadeh M, Hassan ZM, Feizi MA, Najafi F, Hashemi SM. Dendrosomal curcumin significantly suppresses cancer cell proliferation in vitro and in vivo. Int Immunopharmacol. 2012;12(1):226-34. [PubMed ID: 22155627]. https://doi.org/10.1016/j.intimp.2011.11.015.
-
12.
Maheshwari RK, Singh AK, Gaddipati J, Srimal RC. Multiple biological activities of curcumin: a short review. Life Sci. 2006;78(18):2081-7. [PubMed ID: 16413584]. https://doi.org/10.1016/j.lfs.2005.12.007.
-
13.
Suh Y, Afaq F, Johnson JJ, Mukhtar H. A plant flavonoid fisetin induces apoptosis in colon cancer cells by inhibition of COX2 and Wnt/EGFR/NF-kappaB-signaling pathways. Carcinogenesis. 2009;30(2):300-7. [PubMed ID: 19037088]. [PubMed Central ID: PMC2722149]. https://doi.org/10.1093/carcin/bgn269.
-
14.
Anuchapreeda S, Leechanachai P, Smith MM, Ambudkar SV, Limtrakul PN. Modulation of P-glycoprotein expression and function by curcumin in multidrug-resistant human KB cells. Biochem Pharmacol. 2002;64(4):573-82. [PubMed ID: 12167476]. https://doi.org/10.1016/s0006-2952(02)01224-8.
-
15.
Lopes-Rodrigues V, Sousa E, Vasconcelos MH. Curcumin as a modulator of P-glycoprotein in cancer: Challenges and perspectives. Pharmaceuticals (Basel). 2016;9(4). [PubMed ID: 27834897]. [PubMed Central ID: PMC5198046]. https://doi.org/10.3390/ph9040071.
-
16.
Shen L, Ji HF. The pharmacology of curcumin: is it the degradation products? Trends Mol Med. 2012;18(3):138-44. [PubMed ID: 22386732]. https://doi.org/10.1016/j.molmed.2012.01.004.
-
17.
Tahmasebi Birgani M, Erfani-Moghadam V, Babaei E, Najafi F, Zamani M, Shariati M, et al. Dendrosomal nano-curcumin; The novel formulation to improve the anticancer properties of curcumin. Prog Biol Sci. 2015;5(2):143-58. https://doi.org/10.22059/pbs.2015.55525.
-
18.
Zamani M, Sadeghizadeh M, Behmanesh M, Najafi F. Dendrosomal curcumin increases expression of the long non-coding RNA gene MEG3 via up-regulation of epi-miRs in hepatocellular cancer. Phytomedicine. 2015;22(10):961-7. [PubMed ID: 26321746]. https://doi.org/10.1016/j.phymed.2015.05.071.
-
19.
Keshavarz R, Bakhshinejad B, Babashah S, Baghi N, Sadeghizadeh M. Dendrosomal nanocurcumin and p53 overexpression synergistically trigger apoptosis in glioblastoma cells. Iran J Basic Med Sci. 2016;19(12):1353-62. [PubMed ID: 28096969]. [PubMed Central ID: PMC5220242]. https://doi.org/10.22038/ijbms.2016.7923.
-
20.
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402-8. [PubMed ID: 11846609]. https://doi.org/10.1006/meth.2001.1262.
-
21.
Chabner BA, Roberts TJ. Timeline: Chemotherapy and the war on cancer. Nat Rev Cancer. 2005;5(1):65-72. [PubMed ID: 15630416]. https://doi.org/10.1038/nrc1529.
-
22.
Ebrahimi S, Mollaei H, Hoshyar R. Ziziphus Jujube: a review study of its anticancer effects in various tumor models invitro and invivo. Cell Mol Biol (Noisy-le-grand). 2017;63(10):122-7. [PubMed ID: 29096755]. https://doi.org/10.14715/cmb/2017.63.10.19.
-
23.
Mollaei H, Babaei E. Therapeutic Potential of Novel Nano-Based Curcumin Compounds In Vitro and In Vivo. Asian Pac J Cancer Prev. 2017;18(4):885-8. [PubMed ID: 28545183]. [PubMed Central ID: PMC5494236]. https://doi.org/10.22034/APJCP.2017.18.4.885.
-
24.
Strimpakos AS, Sharma RA. Curcumin: preventive and therapeutic properties in laboratory studies and clinical trials. Antioxid Redox Signal. 2008;10(3):511-45. [PubMed ID: 18370854]. https://doi.org/10.1089/ars.2007.1769.
-
25.
Cretu E, Trifan A, Vasincu A, Miron A. Plant-derived anticancer agents - curcumin in cancer prevention and treatment. Rev Med Chir Soc Med Nat Iasi. 2012;116(4):1223-9. [PubMed ID: 23700916].
-
26.
Doello K, Ortiz R, Alvarez PJ, Melguizo C, Cabeza L, Prados J. Latest in Vitro and in Vivo Assay, Clinical Trials and Patents in Cancer Treatment using Curcumin: A Literature Review. Nutr Cancer. 2018;70(4):569-78. [PubMed ID: 29708445]. https://doi.org/10.1080/01635581.2018.1464347.
-
27.
Govindaraju R, Karki R, Chandrashekarappa J, Santhanam M, Shankar AKK, Joshi HK, et al. Enhanced Water Dispersibility of Curcumin Encapsulated in Alginate-Polysorbate 80 Nano Particles and Bioavailability in Healthy Human Volunteers. Pharm Nanotechnol. 2019;7(1):39-56. [PubMed ID: 30666922]. [PubMed Central ID: PMC6691850]. https://doi.org/10.2174/2211738507666190122121242.
-
28.
Montazeri M, Pilehvar-Soltanahmadi Y, Mohaghegh M, Panahi A, Khodi S, Zarghami N, et al. Antiproliferative and Apoptotic Effect of Dendrosomal Curcumin Nanoformulation in P53 Mutant and Wide-Type Cancer Cell Lines. Anticancer Agents Med Chem. 2017;17(5):662-73. [PubMed ID: 27604683]. https://doi.org/10.2174/1871520616666160815124537.
-
29.
Zhao L, Gu Q, Xiang L, Dong X, Li H, Ni J, et al. Curcumin inhibits apoptosis by modulating Bax/Bcl-2 expression and alleviates oxidative stress in testes of streptozotocin-induced diabetic rats. Ther Clin Risk Manag. 2017;13:1099-105. [PubMed ID: 28894373]. [PubMed Central ID: PMC5584885]. https://doi.org/10.2147/TCRM.S141738.
-
30.
Glienke W, Maute L, Wicht J, Bergmann L. Curcumin inhibits constitutive STAT3 phosphorylation in human pancreatic cancer cell lines and downregulation of survivin/BIRC5 gene expression. Cancer Invest. 2010;28(2):166-71. [PubMed ID: 20121547]. https://doi.org/10.3109/07357900903287006.
-
31.
Garg H, Suri P, Gupta JC, Talwar GP, Dubey S. Survivin: a unique target for tumor therapy. Cancer Cell Int. 2016;16:49. [PubMed ID: 27340370]. [PubMed Central ID: PMC4917988]. https://doi.org/10.1186/s12935-016-0326-1.
-
32.
Sah NK, Seniya C. Survivin splice variants and their diagnostic significance. Tumour Biol. 2015;36(9):6623-31. [PubMed ID: 26245993]. https://doi.org/10.1007/s13277-015-3865-5.
-
33.
Malcles MH, Wang HW, Koumi A, Tsai YH, Yu M, Godfrey A, et al. Characterisation of the anti-apoptotic function of survivin-DeltaEx3 during TNFalpha-mediated cell death. Br J Cancer. 2007;96(11):1659-66. [PubMed ID: 17505517]. [PubMed Central ID: PMC2359927]. https://doi.org/10.1038/sj.bjc.6603768.
-
34.
Zhang YJ, Xiang H, Liu JS, Li D, Fang ZY, Zhang H. Study on the mechanism of AMPK signaling pathway and its effect on apoptosis of human hepatocellular carcinoma SMMC-7721 cells by curcumin. Eur Rev Med Pharmacol Sci. 2017;21(5):1144-50. [PubMed ID: 28338175].
-
35.
Chen D, Dai F, Chen Z, Wang S, Cheng X, Sheng Q, et al. Dimethoxy curcumin induces apoptosis by suppressing survivin and inhibits invasion by enhancing e-cadherin in colon cancer cells. Med Sci Monit. 2016;22:3215-22. [PubMed ID: 27614381]. [PubMed Central ID: PMC5021015]. https://doi.org/10.12659/msm.900802.
-
36.
Thomas H, Coley HM. Overcoming multidrug resistance in cancer: an update on the clinical strategy of inhibiting p-glycoprotein. Cancer Control. 2003;10(2):159-65. [PubMed ID: 12712010]. https://doi.org/10.1177/107327480301000207.
-
37.
Punfa W, Yodkeeree S, Pitchakarn P, Ampasavate C, Limtrakul P. Enhancement of cellular uptake and cytotoxicity of curcumin-loaded PLGA nanoparticles by conjugation with anti-P-glycoprotein in drug resistance cancer cells. Acta Pharmacol Sin. 2012;33(6):823-31. [PubMed ID: 22580738]. [PubMed Central ID: PMC4010380]. https://doi.org/10.1038/aps.2012.34.
-
38.
Shakor AB, Atia M, Ismail IA, Alshehri A, El-Refaey H, Kwiatkowska K, et al. Curcumin induces apoptosis of multidrug-resistant human leukemia HL60 cells by complex pathways leading to ceramide accumulation. Biochim Biophys Acta. 2014;1841(12):1672-82. [PubMed ID: 25240837]. https://doi.org/10.1016/j.bbalip.2014.09.006.
-
39.
Yoshimori M, Takada H, Imadome K, Kurata M, Yamamoto K, Koyama T, et al. P-glycoprotein is expressed and causes resistance to chemotherapy in EBV-positive T-cell lymphoproliferative diseases. Cancer Med. 2015;4(10):1494-504. [PubMed ID: 26153782]. [PubMed Central ID: PMC4618620]. https://doi.org/10.1002/cam4.494.