1. Background
Cancer is one of the three most important causes of mortality in humans throughout the world, and colorectal cancer (CRC) is estimated to be amongst the first five frequent cancers in both genders (1). The Lynch syndrome (LS) is the most common form of hereditary cancers, in the context of which the incidence of CRC, uterus cancer, and some other extracolonic cancers such as the stomach, hepatobiliary tract, small intestine, urinary tract, brain, breast, and skin is higher than the average risk of the general population (2). The colorectal cancers associated with LS are usually early-onset and include 3 - 5% of all CRCs (3). Clinical criteria such as Amsterdam I/II (ACI/II) and revised Bethesda guidelines are routinely used to screen the CRC associated with LS, entitled as hereditary nonpolyposis colorectal cancer (HNPCC) (4). The HNPCC phenotype has clinically a heterogeneous pattern encompassing LS (4%), Lynch-like syndrome (< 1%), and familial colorectal cancer type X (FCC-X) (2 - 4% ) (10, 11).
The underlying molecular event in LS mainly involves a germline mutation in DNA mismatch repair genes (MMRs), including MLH1, MSH2, MSH6, and PMS2 (5). Mismatch repair deficiency causes the accumulation of mutations in cancer-related genes, accelerating tumorigenesis events, and leads to a unique molecular phenomenon entitled “microsatellite instability (MSI)”, which can be used as a surrogate marker for MMR deficiency for the molecular screening of LS (6).
Besides LS, about 15% of sporadic CRC patients present MSI-H in their tumors, who are usually affected at a higher age than LS patients (7). The molecular defect in MSI-H sporadic CRC also involves the epigenetic silencing of MLH1 due to promoter hypermethylation (8). According to different studies, a significant portion of these tumors has a specific common mutation in BRAF (V600E), which is not generally found in LS-associated tumors. Therefore, this mutation has been suggested as a surrogate marker to distinguish MSI-H sporadic CRC from LS-associated CRC (8-10).
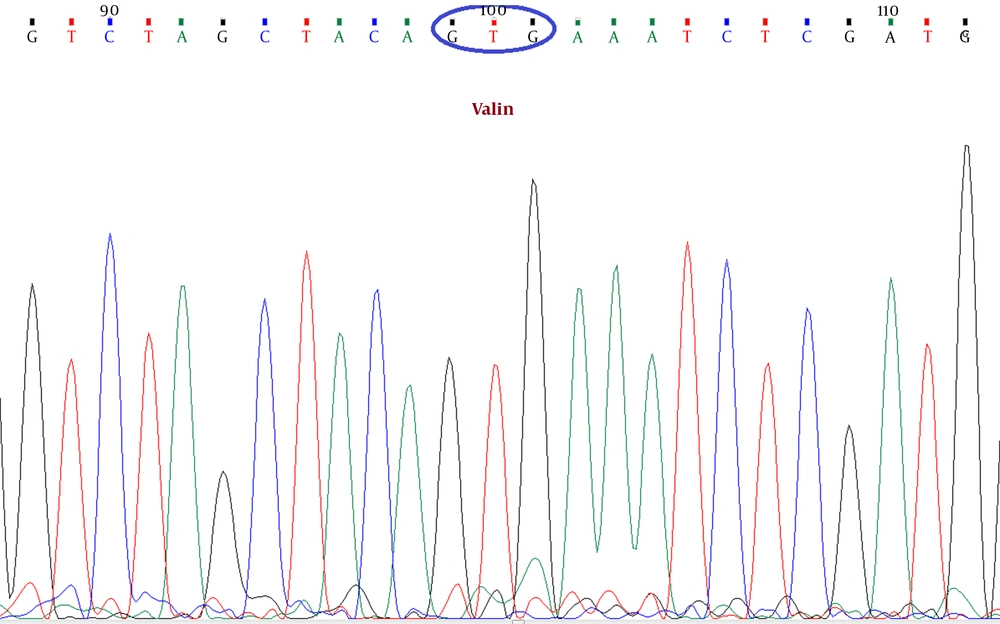
The BRAF gene encodes a cytoplasmic serine-threonine kinase of the Raf family. Gain-of-function mutations in BRAF can lead to the destruction of the kinase domain, which includes a hotspot at the nucleotide position of 1,796, where a T>A substitution leads to an amino acid change from glutamate to valine at the residue 600 (i.e., V600E) (11). This is the most common mutation of BRAF, including up to 90% of all mutations in CRC (12). Oncogenic BRAF mutations have been found in 4 - 12% of all colorectal tumors, regardless of the type or pathologic stage of the tumor (12-14). Several studies have indicated a prognostic value for the BRAF V600E mutation, and mutant CRCs have presented accelerated recurrence and a poor prognosis (15).
Familial-CRC-type-X is characterized by AC positivity and MMR-proficient tumors without mutations in MMR genes (16-18). Meanwhile, some parts of MLH1 defective tumors may present MMR-proficiency in immunohistochemistry (IHC) due to the immunogenicity of the truncated protein (2, 6).
Although FCCX constitutes a major portion of hereditary CRC cases, its molecular features remain poorly defined. More knowledge about the genetics and molecular basis of this subset of hereditary CRC can provide vital hints of disease-predisposing factors and help to develop new efficient early diagnostic and targeted therapeutic strategies (18). Whether or not some FCCX cases harbor sporadic MLH1-defective tumors and MMR-proficiency is uncertain because of the false-negative results of MSI-testing and IHC, and the current study was designed to divulge this issue. Among Iranian FCCX patients with different clinicopathologic features (2, 16), we evaluated the presence of the V600E BRAF mutation as a known predictive and prognostic marker in CRC and a well-defined surrogate for detecting sporadic MLH-defective CRCs.
2. Methods
2.1. Patients and Samples
This was a descriptive study to reveal more clearly a molecular feature of FCCX in central Iran, Isfahan. Among 2,460 CRC patients registered in Poursina Hakim Digestive Diseases Research Center and Iranian Cancer Control Charity Institute, two referral centers for cancer patients in Isfahan, 48 families with FCCX were screened for MMR deficiency using IHC-MMRs staining and MSI testing according to ACII, which included the presence of at least three members with one of LS-associated cancers (colorectal, uterus, gastrointestinal, renal, breast, brain, skin, and pelvic) in at least two successive generations, as well as a first-degree relationship between at least two out of three affected members and at least one being diagnosed before the age of 50 years (19).
An informed consent form was signed by the patient or a family member. Ethical approval was obtained from the Research Ethics Committee of Shahrekord University of Medical Sciences (ethical ID: 93-6-6). The study received grant number of 1686.
2.2. BRAF Mutation Screening
In this study, a primer pair was designed for the exon 15 of the BRAF gene using the Primer-Blast tool of NCBI (forward primer: ATTGCACGACAGACTGCACA and reverse primer: CTGATGGGACCCACTCCATC) to amplify a DNA sequence enclosing the V600E BRAF mutation. DNA extraction was performed on tumor tissue samples from formalin-fixed paraffin-embedded (FFPE) blocks (20). After PCR, according to standard procedures, Sanger sequencing was done by an ABI PRISM 3100 Genetic Analyzer system. Using Chromas 2.6.4 DNA sequence editing software (Technelysium Pty Ltd), the reported electropherograms were analyzed, and the presence of the V600E BRAF mutation was assessed (Figure 1).
3. Results
Overall, 48 cases (1.95%), out of 2,460 patients with CRC, were finally diagnosed as FCCX. None of the MMR-proficient probands in the 48 studied FCCX families presented the V600E BRAF mutation in tumors’ extracted DNA. According to pedigrees, affected members in MMR-proficient CRC families had an average age of 51.7 years at diagnosis. The frequencies of right-sided and left-sided colorectal tumors were 20.8 and 79.2%, respectively. The number of cancer patients in the 48 identified families with FCCX was 286, and CRC, gastric, and lung cancers with 39.9, 10.1, and 8.4%, respectively, were the most frequent cancers (Table 1).
| Tumor location | No. (%) |
|---|---|
| Colorectal | 114 (39.9) |
| Stomach | 29 (10.1) |
| Lung | 24 (8.4) |
| Breast | 23 (8.0) |
| Brain | 18 (6.3) |
| Hepatobiliary tract | 14 (4.9) |
| Intestine | 12 (4.2) |
| Prostate | 8 (2.8) |
| Uterus | 8 (2.8) |
| Skin | 6 (2.1) |
| Hematopoietic system | 6 (2.1) |
| Bladder | 6 (2.1) |
| Thyroid | 4 (1.4) |
| Testis | 4 (1.4) |
| Bone | 4 (1.4) |
| Kidney | 2 (0.7) |
| Pancreas | 2 (0.7) |
| Nasopharynx | 2 (0.7) |
| Total | 286 (100) |
Overall, 16/48 of FCCX probands (33.3%) were pathologically diagnosed at early stages (I or II TNM stage). In addition, the mortality rate of cancer among the FCCX probands was 22/48 (45.8%).
4. Discussion
In this study, the V600E BRAF mutation was evaluated in 48 FCCX probands. The hot-spot V600E BRAF mutation can be applied as a prognostic factor and a predictive marker for anti-EGFR targeted-therapy in CRC. It is also used to distinguish sporadic CRC from familial CRC and as a surrogate marker for the evaluation of MMR deficiency due to MLH1 promoter hypermethylation, an event happening in an average of 15% of CRCs (21). Moreover, differences in the clinicopathologic features of familial CRCs between Iranians and other populations can be related to different genomic structures and molecular pathways involved in tumorigenesis (6, 22, 23).
4.1. The Prevalence of BRAF Mutation in Tumor DNA
In this study, none of the 48 assessed probands presented with the V600E BRAF mutation, as evidenced by sequencing tumor extracted DNA samples. The frequency of this mutation in CRC tumors has been variable in different studies, ranging from 1 to 22% and from a lower prevalence in some Asian populations to a higher prevalence in some western nations (24-28). Meanwhile, the frequency of the V600E BRAF mutation in early-onset right-sided MSI-H CRC has been reported to be higher than in other CRC types (26-28). For example, despite the low frequency of the mutation in unselected Taiwanian CRC patients (about 1%) (25), this mutation was found in 19% (11 of 59 cases) of Taiwanian patients with early-onset CRC (26). There are limited surveys on the prevalence of the V600E BRAF mutation in patients with familial CRC. In a Swedish study, 194 CRC patients with a positive family history of the disease were investigated, 26 (13.4%) of whom revealed the V600E BRAF mutation in their tumors; however, hereditary syndromes such as familial adenomatous polyposis (FAP) and LS had been excluded from this study (11).
Based on different studies, it seems that the prevalence of the V600E BRAF mutation in CRC tumor tissues is lower in the West and South of Asia compared to Western populations (25, 29, 30). For example, the prevalence of this mutation has been estimated 2.5 and 6.7% in two studies performed in Saudia Arabia and Turkey, respectively (25, 29).
There is insufficient data on the prevalence of the V600E mutation among Iranians; meanwhile, given the presence of several ethnicities in Iran, the prevalence of this mutation may vary in different areas of the country. In an Iranian study performed in the southwest of the country, 37 of 80 (46.2%) non-selective CRC cases had a heterozygous genotype for the V600E BRAF mutation (30). In another study in the northwest of Iran, the mutation was found in none of 30 non-selective CRC cases (31). Nevertheless, no survey has been done yet on Iranians with familial CRC in terms of V600E BRAF mutation prevalence. Given the low sample size of the present study, more evaluations on other Iranian populations with larger numbers of patients with familial CRC can deliver a more accurate estimation of V600E BRAF mutation prevalence.
4.2. Clinicopathologic Features
Most CRC tumors with mutant BRAF, according to different studies, present characteristic clinicopathological features such as older age at diagnosis, right-sided location, and an advanced pathologic stage at diagnosis (32). According to our findings, about one-third of the cases studied here were diagnosed at early pathological stages (stage I and II), and most of them were identified at advanced stages. Given that all of the samples showed the wild-type BRAF, the identification of the patients at more advanced stages may be due to delayed diagnosis because of the lack of active CRC screening programs in Iran. Moreover, just one-fifth of the tumors were located at the right side of the colon, and most of them were left-sided. This feature is compatible with non-mutant BRAF tumors, an issue that was approved by our findings.
4.3. Predictive Value of BRAF Mutation
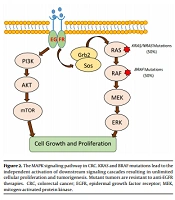
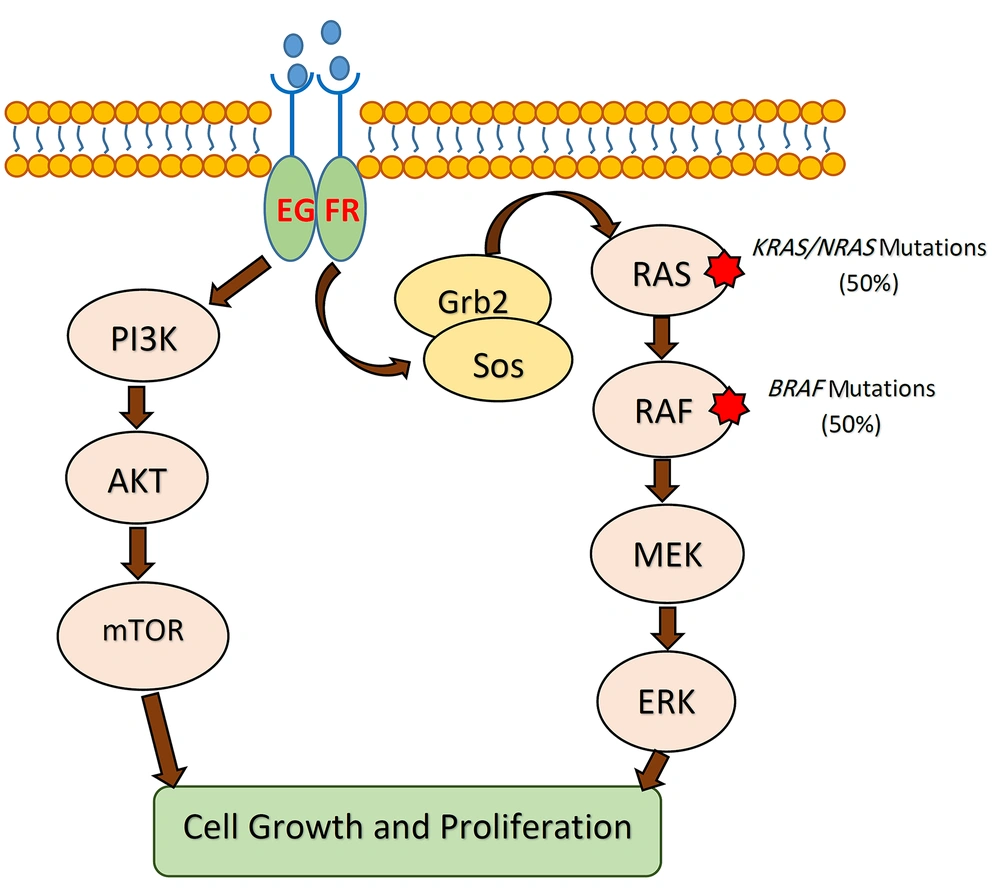
According to several studies, tumor DNA analysis for BRAF and KRAS mutations can help clinicians to choose a cost-effective and efficient drug to treat the patient by targeted therapy (33). The BRAF and KRAS are effector proteins in the mitogen-activated protein kinase (MAPK) pathway, which is triggered by ligand-bound epidermal growth factor receptor (EGFR) (34). The monoclonal antibodies (mAbs) manufactured against EGFR are now used as key drugs in cancer targeted-therapy to treat metastatic CRCs (35). Gain-of-function mutations in both oncogenes (i.e., BRAF and KRAS) can be used as strong predictors of resistance to targeted anti-EGFR therapy (36, 37). Accordingly, BRAF mutations can be used as predictors of the therapeutic response, particularly in metastatic CRCs (Figure 2). Given that wild-type BRAF was identified in all tumor DNA samples in our study, targeted therapy with anti-EGFR agents could be considered for the patients.
The MAPK signaling pathway in CRC. KRAS and BRAF mutations lead to the independent activation of downstream signaling cascades resulting in unlimited cellular proliferation and tumorigenesis. Mutant tumors are resistant to anti-EGFR therapies. CRC, colorectal cancer; EGFR, epidermal growth factor receptor; MEK, mitogen-activated protein kinase.
4.4. Prognostic Value of BRAF Mutation
As mentioned, the BRAF V600E mutation is a prognostic marker in CRC, predicting a higher recurrence rate and a shorter survival (15). According to our results, two-thirds of the patients had been diagnosed at late stages, and nearly half of them were deceased at the time of the study. Nevertheless, regarding the lack of mutant BRAF in all the studied probands, late diagnosis could justify the relatively high mortality rate in our patients. So, the survival of patients would be improved through implementing regular screening programs, an essential health-related process which is currently not performed in Iran.