Abstract
Background:
In vitro biofilm formation of H. pylori is demonstrated; however, its potential role in the persistent infection of the human stomach has not yet been addressed.Objectives:
The aim of this study was to assess the biofilm formation of clinical H. pylori isolates on an epithelial cell line, a line that produces mucin.Methods:
H. pylori isolates consisting of an efficient (19B) and a weak (4B) biofilm formation ability, were selected from screening of the clinical isolates. Their adhesion index was determined after 2h incubation with the semi-confluent monolayers of MKN-45 cells. Their biofilm formation was evaluated after 24 and 72 h incubation with MKN-45 cells using a modified adherence assay developed in this work. Production of biofilm was quantitatively assessed by CFU enumeration and qualitatively by the immunofluorescence, and scanning-electron-microscopic (SEM) methods. Due to the importance of mucin in the binding of H. pylori and biofilm formation, the binding strength of the mucin binding protein, MUC5AC, and MUC1 with docking was investigated using cluspro webserver.Results:
Using MKN-45 epithelial cell line as a model, significant differences were observed between the adhesion index of 19B and 4B isolates. After 24h, both isolates were able to form biofilms with significantly higher numbers of CFU for the 19B isolate. These results were confirmed by immunofluorescence and SEM such that after 24h, a cluster of coccoid bacteria on the MKN-45 cells in the form of microcolonies was observed. The docking results showed that MUC5AC demonstrated the most favorable interaction with H. pylori urease and BabA with docking energy scores of -931.1 and -906.3 kcal.mol-1, respectively.Conclusions:
By developing an appropriate in situ biofilm assay, we investigated biofilm formation by clinical H. pylori isolates on the MKN-45 epithelial cell line. The establishment of such an in-situ model for studying the biofilm formation ability of clinical isolates can also be used to study cell-bacteria interactions in the context of a complex biofilm and also as a model for drug screening applications.Keywords
Helicobacter pylori MKN45 Cell Line Adherence Biofilm Coccoid Form
1. Background
Over one half of the world’s population is colonized with Helicobacter pylori. Hence it is considered as one of the most frequent agents of human infections (1).
Adhesion to gastric cells is a crucial step in the formation of a successful infection because it provides protection from natural clearance mechanisms (2). In spite of the fact that many investigations have been performed to understand the role of virulence factors in favoring the more severe outcome of H. pylori infection, it is not well known why this infection still persists in the absence of fatal outcome for years and lead to a chronic infection.
The first study investigating in vitro biofilm formation by H. pylori corresponds to the work of Stark et al. (3), which was followed by other investigators (4-8). Very few in vivo studies have been undertaken, and they are limited to the observation of biopsy specimens from the patients’ stomach (9, 10). In light of growing evidence, revealing the impacts of biofilm on antibiotic action and host immune responses, the importance of the biofilm formation ability of H. pylori is increasingly being recognized.
In prior studies, the ability of different H. pylori strains in vitro models on an abiotic surface such as polystyrene or glass has approved the biofilm formation ability of this bacterium, but those surfaces did not mimic the real state of stomach surface. Using the C57BL/6J mice model, we have previously studied the ability of H. pylori to form typical biofilms in the mouse stomach mucosa, but animal models have some limitation such as cost, ethical rules, and hard to set up (11, 12). Multiple investigators have studied the advantages of an in-situ system for studying biofilm formation by bacteria. In a recent report, Salas-Jara et al. (13) have studied the ability of Lactobacillus fermentum UCO-979C to form biofilm on cell line. The cell line culture method was also used as model for human skin wound. Using histological assessment, wide-field fluorescence microscopy, stereo-fluorescence microscopy, and SEM, Ashrafi et al. (14), showed the biofilm formation on cell lines.
In vitro evaluation of H. pylori adherence to epithelial cell models using microtiter-plates and adherence assessment by calculating the adhesion index are now well defined. However, these tests can only demonstrate the interaction of the bacteria with the host cells under a short period and are not sufficient to demonstrate the biofilm formation.
2. Objectives
The objective of present study was to assess the biofilm formation of clinical isolates of H. pylori on the human epithelial cell model MKN-45 cell-line, a line that produces the mucin. For this purpose, an in-situ assay was developed with two advantages: first, to permit development of a biofilm in-situ; second, the presence of mucin, which is present in real conditions in the stomach and gastrointestinal tract. As mucin could play a critical role in the colonization of H. pylori, in this study, we investigated the interaction of adhesive factors with mucin using bioinformatics tools.
3. Methods
3.1. Bacterial Isolates and Growth Conditions
A collection of thirty clinical H. pylori isolates, isolated from patients showing signs of chronic gastritis, were taken with informed consent for this study. Isolates were cultured onto brucella agar base (Merck, Germany), supplemented with 5% defibrinated sheep blood and antibiotics at 37°C under microaerophilic atmosphere for 3 - 7 days.
The isolate were identified by Gram staining, biochemical tests, including catalase, oxidase, urease, and nitrate. As previously described, the molecular identity of H. pylori strains was confirmed by PCR amplification of gene 16sRNA and glm gene (glm-F5’GGATAAGCTTTTAGGGGTGTTAGGGG3’, glm-R5’ GCATTCACAAACTTATCCCCAATC 3’, 16s rRNA-F ‘GAAGATAATGACGGTATCTAAC 3’, 16s rRNA-R5’ ATTTCACACCTGACTGACTAT 3’) (15, 16).
3.2. In Vitro Screening of Biofilm Formation
It was done by a method adapted from a previously described protocol, with some modifications (7, 17, 18). The grown colonies were harvested from culture-plates and inoculated into brucella broth (Biolife, Italy) supplemented with 2% (w/v) fetal calf serum (FCS) and 0.3% (w/v) glucose (Merck, Germany), and incubated overnight at 37°C in the microaerophilic atmosphere with shaking at 100 rpm. An optical density of broth cultures adjusted to 0.2 at 600 nm (A600), equivalent to 5 - 8 × 103 CFU/mL, was inoculated (250 µL) into the 96-well flat-bottomed culture plates (BIOFIL, Jet Bio-Filtration Products Co., China) and incubated at 37°C under the microaerophilic condition for six days. After biomass formation, non-adherent bacteria were removed by washing (3×) with sterile PBS. The tightly attached bacteria were fixed with 99% ethanol (200 µL per well) for 20 min and air-dried. Plates were then stained with 1% crystal violet (200 µL per well) for 5 min, and the excess stain was rinsed away with running tap water. The dried plates were treated with 33% (v/v) glacial acetic acid (160 µL per well), and their optical density (OD) was measured at 505 nm by ELISA reader (SCO, Germany). The following criteria were used for biofilm classification in H. pylori clinical isolates: OD ≤ ODcontrol = Non-biofilm-former, ODcontrol < OD ≤ 2 × ODcontrol = Weak biofilm-former, 2 × ODcontrol < OD ≤ 4 × ODcontrol = Moderate biofilm former , OD > 4 × ODcontrol = Strong biofilm-former (19).
3.3. Epithelial Cell Line and Monolayers Preparation
The MKN-45 (IBRC C10137) cell line (Iranian Biological Recourse Center: IBRC) was routinely grown in RPMI1640 + 20% fetal bovine serum (FBS: Gibco, USA) and antibiotics including 40 µg/mL gentamicin (sigma - USA) and 2.5 µg amphotericin B per mL of media (Cipia, India).
3.4. Adherence Assay
The confluent cell culture was trypsinized, adjusted to 4 -5 × 105 cells/ml, transferred into 12-well tissue culture plates (Orange Scientific), and incubated until approximately 106 cells/well. H. pylori 19B and 4B Isolates were grown for three days, and the negative control, E. coli ATCC 2995 isolate, was grown on LB agar plate overnight. Inoculums (× 109 bacteria/mL) were prepared in PBS; their number was determined by the CFU method. Before inoculation, monolayers were washed and incubated with 0.9 mL RPMI medium + 3% FBS. The cells in the wells were counted and inoculated with the bacterial suspension at a ratio of 100 bacteria/host-cell (20) and incubated for 2 h. Then, the monolayers were vigorously washed (3×), and the cells (3 wells) were lysed by addition of 1 mL of deionized sterile water and incubated at 37°C for 30 min. The resulting suspension was used for enumeration of adherent bacteria by CFU method, and to obtain the adhesion index, three wells were trypsinized for enumeration of host cells by direct counting in a haemocytometer. The adhesion index was defined as the mean number of adhering bacteria per cell, which was obtained from at least three independent assays (21).
3.5. Evaluation of Biofilm Formation
Six-well culture plates were seeded with MKN-45 cells (6 - 7 × 105 cells/well) in RPMI 1640 containing 20 % FBS and were grown for 3 - 7 days to 80% confluence by renewing their medium every day. H. pylori cultures were harvested into F-12 Ham nutrient medium (Sigma-Aldrich, USA) washed two times and adjusted to 109 bacteria/mL. Prior to bacterial-inoculation, medium was replaced with F-12 Ham medium supplemented with 5% FBS and 2.5 µg/mL of amphotericin B, then inoculated with bacterial-suspension and incubated for 3 h. The wells were drained off, washed to eliminate planktonic bacteria, and incubated in fresh medium for 24 and 72 h, during which the medium was renewed two times a day. After each period, the wells were washed vigorously (3×), then treated with 0.1% Triton X-100 in PBS for 10 min to lyse the host-cells and disperse the biofilm. Serial dilutions of the lysate were used for enumeration of bacteria by the CFU method.
3.6. Biofilm Monitoring by Immunofluorescence Staining
Inoculated host-cells grown on microscopic chamber slides were washed 3× after incubation period (to eliminate planktonic-bacteria). Cells were fixed with 4% paraformaldehyde (25°C for 10 min), washed (3×) for 5 min with PBS-T (PBS- 0.05%, v/v Tween-20) to eliminate fixative traces and to permeabilize the cells. Cells were blocked with 3% BSA in PBS-T (1 h, 25°C), washed in PBS-T for 5 min and incubated with the diluted antibody (1:1000 in PBS-T + 1% BSA) of a home-made anti H. pylori antibody at 25°C for 1 h (22). They were washed (3×) with PBS-T (10 min) and incubated (1 h) with the goat anti-rabbit IgG conjugated to rhodamine in PBS-T + 1% BSA (Sigma, USA) with a concentration of 0.01 µg/mL, washed with PBS-T (3×) for 10 min. Also, nuclear staining was performed as follows: cells were stained with DAPI (4’, 6-diamidino-2-phenylindole, Roche, Germany), diluted 1:1000 in PBS-T or PI (Propidium iodide, Molecular Probes) for 5 min, and then rinsed with PBS for another 5 min (23).
3.7. Biofilm Monitoring by Scanning Electron Microscopy
Samples were fixed in 2.5% glutaraldehyde for 30 min, dehydrated with increasing concentrations of ethanol (30, 50, 70, 90) and 100% (2×) each for 5 min. Samples were stored in desiccators until they were coated with gold-palladium sputter for two 200-seconds intervals (Nano Structured coating Co. Iran). Biofilm monitoring by scanning electron microscopy (SEM) micrographs were performed using TESCAN VEGA3S electron microscope at 30 KV (10).
3.8. Mucin and Mucus-Binding Proteins of H. pylori Interaction
In-silico interaction of mucus-binding proteins of H. pylori with mucins was assessed using Cluspro protein-protein Docking (24-27) which, is the fully automated, web-based program for the computational docking of protein structures. The docking algorithms evaluate billions of putative complexes; a filtering method by selecting those with good electrostatic and desolvation free energies for further clustering was applied. According to literature, MUC5AC and MUC1 were analyzed for their in-silico interaction with Lewis b antigen binding by the H. pylori-BabA and urease.
The three-dimensional structure of mucin-binding proteins and mucins were downloaded from in the Protein Data Bank (PDB) database (https: //www.rcsb.org/). 1e9z (crystal structure of H. pylori urease), 4zh7 (Lewisb antigen binding by the H. pylori- BabA), 5ajn (MUC5AC), 2acm (human mucin 1 (MUC1)), were used for docking analysis after removing solvent and ligands with UCSF Chimera (28). Individual docking procedures have been performed for each ligand-protein complex. The findings have been ranked in the order of rising docking energies which the lowest binding energy of each cluster was considered. Docked complexes were further analyzed by using UCSF Chimera visualization.
3.9 Statistics
Standard statistical analysis by Graph Pad Prism 7 was used to evaluate the reproducibility of the test in various wells (standard deviations) and to determine the significant difference (P-value) between the adhesion index of 19B and 4B H. pylori isolates as well as ATCC 2995 E. coli isolate.
4. Results
4.1. Isolates Screening
All 30 isolates were able to form biofilm on polystyrene microplates, with differences between the amounts of biofilm among them. According to our result, 3.3% of isolates are strong biofilm former, 10% were weak biofilm former, and the rest were moderate biofilm former. Two isolates with the highest and lowest ability in biofilm formation, named (19B and 4B), were selected for further in-situ biofilm formation analysis on the epithelial cell line.
4.2. Adherence to MKN-45 cells
Evaluation of adhesion index after 2 h of incubation revealed that both isolates were able to adhere to MKN-45 cells. However, their adhesion indices were significantly different and were 12.9 and 9.6 for 19B and 4B, respectively. Negative-control, 2995 E. coli strain, could not adhere to MKN-45 cells.
4.3. Quantitative Evaluation of Biofilms
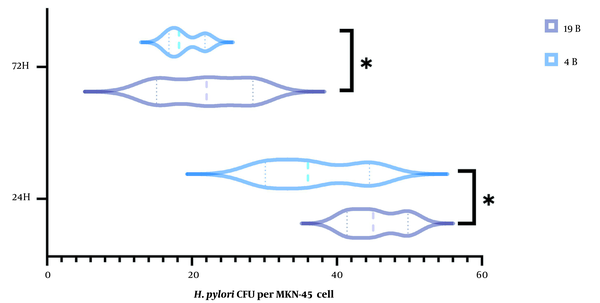
The number of bacteria recovered from lysed cells after 24 and 72 h incubation was counted. H. pylori isolate 19B produced significantly higher amount of biofilm on the host cells compared to the 4B isolate (Figure 1). The number of bacteria reduced by two folds in both isolates after 72 h.
Vilion plot of bacterial enumeration in the H. pylori biofilms after 24 and 72 h.
4.4. Inverted Light Microscope Observation
Both spindle-shaped and oval cells growing in monolayers and single round cells or clumps in suspension were seen in inverted light microscope images (Appendix 1 in Supplementary File).
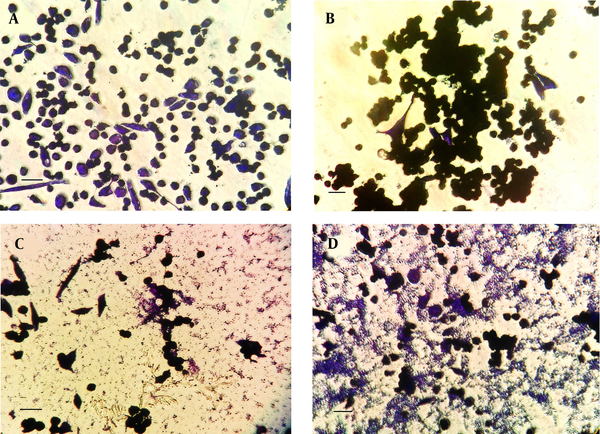
Microscopic analysis of crystal-violet-stained H. pylori isolate 19B biofilms on the MKN-45 cells indicated that cells aggregated together to form microcolonies (Figure 2). After 24 h, cells had begun to aggregate and form small groups (Figure 2B), and by 72 h, larger groups of microcolonies were evident (Figure 2C and D)
Representative inverted micrographs of biofilms formed by clinical isolate H. pylori isolate 19B on MKN-45 cell monolayer after staining with crystal violet for three minutes. Cell cluster morphology (microcolony) strongly suggestive of biofilm formation. A, Control group; B after 24 h; C and D, After 72 h (Scale bar, 200 µm).
4.5. Immunofluorescence Observation of the Biofilms
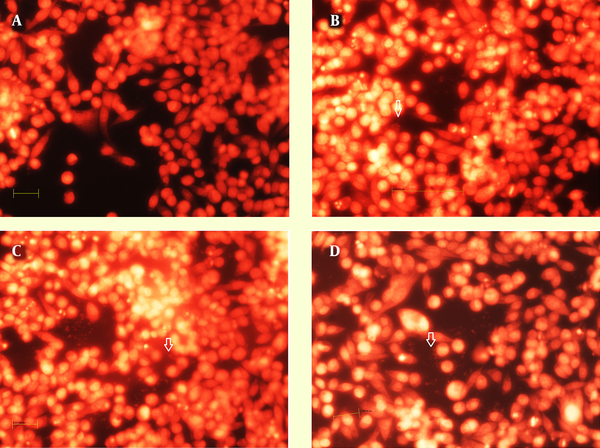
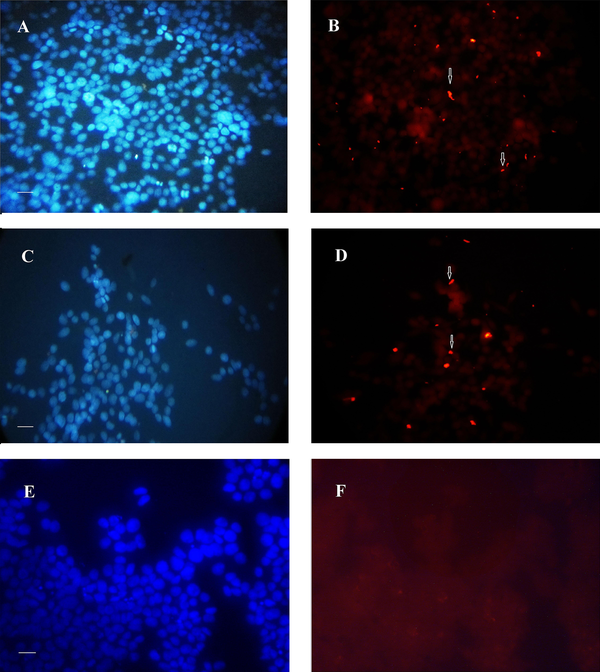
Figure 3 shows the nuclear staining of the cells with propidium iodide but revealed no difference in the number of host cells between the control (Figure 3A) and those infected by the 19B (Figure 3B-C) and 4B isolates (Figure 3D).
Immunofluorescence image of biofilm formed after 24 h by 19B and 4B H. pylori on the MKN-45 cells with nuclei stained with propidium iodide. A, Control without H. pylori inoculation; B-C and D, Incubation with19B and 4B isolates, respectively. Arrowhead shows biofilm formation over MKN-45 monolayer.
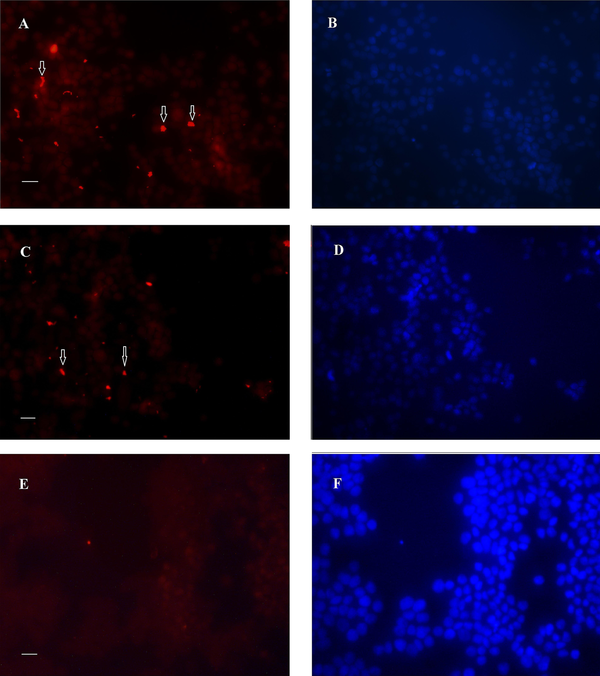
Analysis of the immunofluorescence images after 24 h revealed the clusters of bacteria on the MKN-45 cells for both 19B and 4B isolates, with higher densities for the 19B isolate (Figures 4-5). After 72 h, the same clusters of bacteria were observed embedded in the extracellular matrix.
Immunofluorescence image of biofilms formed after 24 h by 19B and 4B H. pylori isolates on MKN-45 cells. A-B, Rhodamin stained control group visualized by red filter and nuclei stained by DAPI visualized by UV filter, respectively; C and D, Cells inoculated with 19B isolates stained with rhodamin and nuclei stained by DAPI; E and F, Cells inoculated with 4B isolate stained with rhodamin and nuclei stained by DAPI (Scale bar, 200 µm).
Immunofluorescence image of biofilm formed after 72h by 19B and 4B H. pylori on the MKN-45 cells. A-B: control cell nuclei stained with DAPI. 72 hours-biofilm formed on MKN-45 cell monolayer, A, C, and E, Nuclear staining with DAPI; B, D, and F, Immunofluorescence staining with rhodamin conjugated to anti rabbit antibody. Fluorescent bacteria are seen on or between MKN-45 cells; A and B, Control group; C, D, E, and F, Cell monolayer inoculated with H. pylori isolate 19B and 4B, respectively (Scale bar, 200 µm).
4.6. SEM Analysis of Biofilms
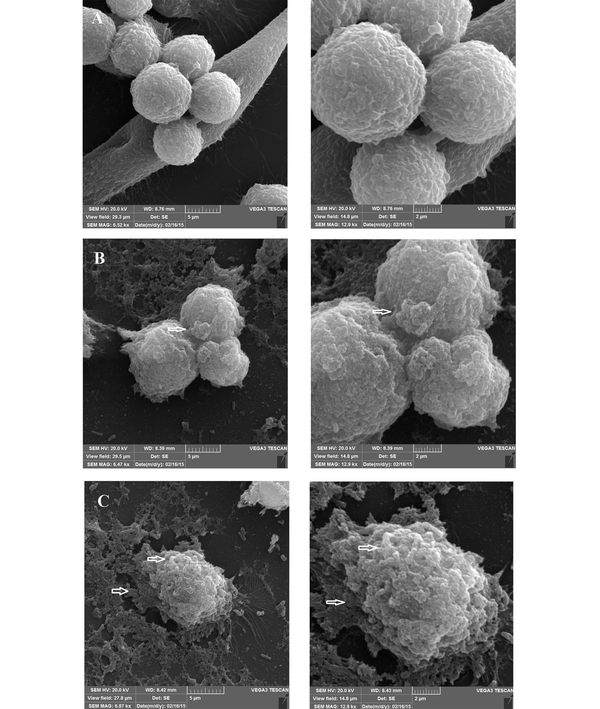
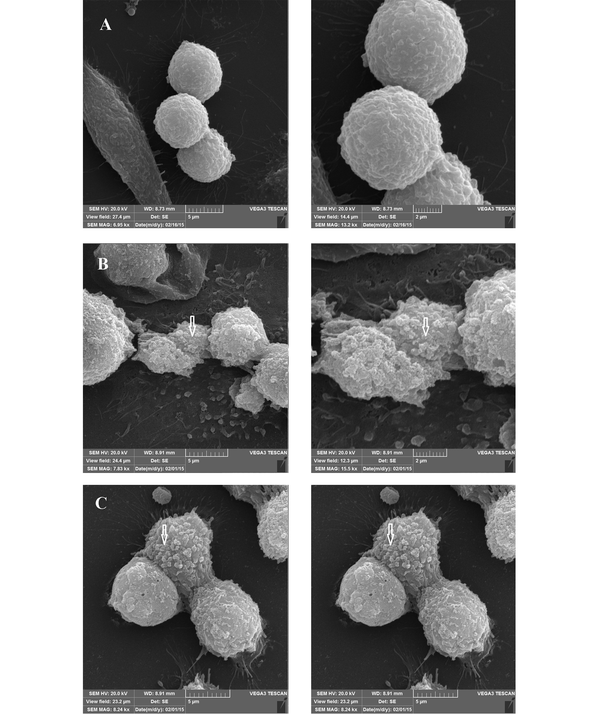
Observation of SEM images 24 h after incubation showed the cell cluster morphology of the cell-associated bacteria, in the form of microcolonies in which the bacterial cells were tightly associated (Figure 6). After 72 h, although the same microcolonies were detectable, a change in the morphology of MKN-45 cells was also observed, especially in the case of those associated with the 19B isolate (Figure 7).
Representative SEM micrographs of 72 hour-biofilms formed by clinical isolate H. pylori on MKN-45 cell monolayer. Scanning electron micrographs showing cell cluster morphology (microcolony) strongly suggestive of biofilm formation. A, Control group; B and C, Cell monolayer inoculated with H. pylori isolate 4B and 19B, respectively.
Representative SEM micrographs of the MKN-45 cells infected by 19B and 4B isolates after 72 h. A, Control; B and C, Infected by 19B and 4B isolates, respectively.
4.7. Docking Analyzing Mucus Binding Proteins with Mucin
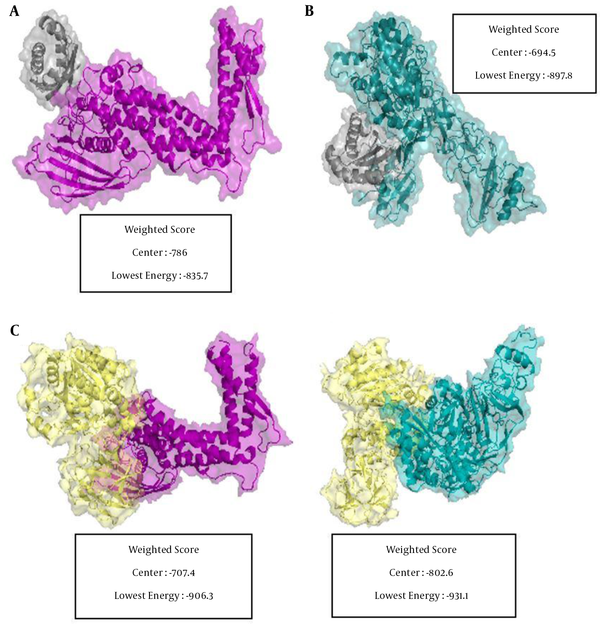
The in-silico docking results between the H. pylori mucin-binding protein and mucin (MUC1 and MUC5AC) revealed that MUC5AC has the highest binding affinity in terms of the protein-protein interaction. Out of the four possible interactions, MUC5AC demonstrated the most favorable interaction with H. pylori urease and BabA with a docking energy score of -931.1 and -906.3 kcal.mol-1, respectively (Figure 8).
Positions and interactions of MUC5AC and MUC1 with H. pylori BabA and urease. A, MUC1 as receptor and BabA as ligand; B, MUC1 as receptor and urease as ligand; C, MUC5AC as receptor and BabA as ligand; D, MUC5AC as receptor and urease as ligand. BabA, urease, MUC5AC, and MUC1 are depicted in purple, yellow, cyan, and gray, respectively. The docking score represents for each part.
5. Discussion
Despite the fact that numerous studies confirmed biofilm formation by H. pylori on non-living environments, a few studies have been shown biofilm formation on humans stomach or other in vivo models. Recently, research has suggested the correlation between H. pylori biofilm and gastric cancer, but more evidence is required to confirm this matter (29). Nowadays, cell culture is considered as a rapid and inexpensive method for investigating the efficacy of drug candidates (30). Therefore, creating an in vivo model for biofilm assay will be useful for studying the relationship between biofilm and cancer and examining antibiotic resistance.
We developed an assay for exploring the biofilm formation by H. pylori via modifying the universal adherence test to evaluate biofilm formation on a host cell model. Comparison of adherence index after 2 h, and evaluation of bacterial count after 24 h, showed a significant difference between isolates 19B and 4B (Figure 1). Thus, despite the suggestion that the 19B isolate has a higher capacity to proliferate on the host cells, a lower number of bacteria was detected after 72 h in both isolates, which might be due to the limitation of host-cell growth conditions and/or presence of non-viable bacteria.
Nuclear staining of the cells after 24 h showed no difference in the number of host cells between control and those infected by 19B and 4B isolates, suggesting that the host cells maintain their integrity after 24 h in all cases (Figure 3). However, evaluation of immunofluorescent images revealed a dense presence of fluorescent stained bacteria, which was higher for the 19B isolate (Figures 4-5).
Inconsistent with our result, Cai et al. (31) used AGS cell line culture to study H. pylori biofilm formation and examined the biofilm formation with a fluorescent microscope (live/Dead staining) but did not elaborate on their work.
By SEM evaluation, the communities of coccoid bacteria were observable on the host cells, for both isolates (Figures 6-7). The presence of bacterial cells on the extracellular matrix was also visible, especially after 72 h. Despite the persistence of microcolonies after 72 h of incubation, signs of host cell deterioration were noticeable in the SEM images, which might explain the decline in bacterial count after 72 h. Previous studies have proposed that some H. pylori isolates are able to form a biofilm in laboratory in the form of coccoids (4).
Hathroubi et al. (32) used a method similar to ours. Additionally, they noticed in SEM images H. pylori totals were not homogeneously dispersed over the outside of the AGS cells yet discovered to a great extent in the recessed cell-cell intersection regions, as revealed previously. This non-uniformity can be seen to some extent in the images of this study (33).
Although several studies have indicated that biofilm formation plays an important role in the pathogenesis of chronic infection, these studies have been performed in vitro using solid surfaces such as glass or plastic, where it is not possible to completely imitate the real conditions of the stomach (4-6, 34).
Goblet cell increased release of secreted mucins to mucosal infection (35). Mucins are secreted cell surface glycoproteins which that some pathogenic bacteria have mechanisms to target specific surface mucins on the other hand; they act as inhibitors of the many bacterial receptors. It is also known that the adherence of H. pylori to the gastric mucosal surface constitutes a critical step in interactions with the host gastric cells and the first step in biofilm formation. Furthermore, it was known that the blood group antigen binding adhesin (BabA) of H. pylori, binds to Lewis b antigen. Gastric mucous layer gel, mainly consisting of the MUC5AC that, harbors glycan-rich domains presenting the Lewis b antigen, which is the most important carrier of the LeB carbohydrate structure in normal gastric tissue (36, 37).
In addition to MUC5AC, MUC1 is the main mucin genes expressed in surface/foveolar epithelial cells in the normal stomach. H. pylori regulate mucin gene expression at the transcriptional level in gastric cells line (36). MUC1 is aberrantly overexpressed by more than 50% of stomach cancers. Still, its role in carcinogenesis remains to be defined, H. pylori upregulate MUC1 expression in gastric cancer cells (38).
Higher mucin expression in the gastric epithelium of H. pylori positive patients than in healthy controls was demonstrated (39). It was shown that in oral cavity H. pylori using BabA, SabA binds to salivary mucins in saliva, and it seems that affect that colonization in various niches along the orogastric infection route and reinfection (40). In another study, it was shown that urease from H. pylori increases the expression of mucin gene including MUC5AC in AGS cell line, so docking urease with mucin was also investigated (36). In conclusion, the interplay between H. pylori infection and mucin secretion is important for attachment. H. pylori bind to gastric mucin. On the other hand, H. pylori infection increases its mucin expression. Therefore, mucin plays an important role in simulating the actual condition of the stomach and is important in studies related to H. pylori binding.
Since the presence of H. pylori has been reported in the subgingival biofilm, due to the importance of mucin in the binding of H. pylori, which is the first step in biofilm formation, this study, we also examined the binding strength by docking methods. As our results showed, H. pylori has the ability to bind to mucin, especially MUC5AC 5AC. Therefore, if the mucin is present in biofilm formation assay, the results of study will be close to the natural conditions.
Matsuda et al. compare gene expression alteration after H. pylori infection in three cell lines, H. pylori infection, AGS, KATO III, and MKN45. They showed that H. pylori cells alter the expression of the transcription factors mRNA such as az, MUCs mRNA in H. pylori-infected cells, in a pattern common to cells. In AGS cells, the Intestinal phenotype is predominated, whereas the gastric phenotype predominated in MKN45 and KATO III cells. In MKN45 cells, H. pylori-induced the three MUCs mRNAs expression (41). Therefore, due to the fact that the expression pattern of mucins in MKN45 cell line is closer to the conditions of the stomach, this cell line was selected to study biofilm formation.
Cole et al. (5) showed that 10% mucin increased planktonic cells, but as they mentioned in that article, they autoclaved mucin with a medium before assay as mucins are glycoproteins probably lose native 3D structure and may serve as a carbon source.
In contrast to this study, we showed that mucin was significantly effective in biofilm formation in our previous study. In our study, we used mucin from the porcine stomach (type II), which contains MUC2 (16). Therefore, it seems that the type of mucin and assessment method affect the results. More studies are needed to reach a definitive conclusion in this regard.
In the current examination, we attempted to emulate, to some extent, the real conditions of the stomach since the cell-line used in this work is able to produce a mucus-layer (42), which would be an advantage for this model to initiate bacterial attachment. Thus, to our knowledge, the present work is the first study, which evaluated the ability of clinical isolates of H. pylori to adhere and form a biofilm in an experimental human epithelial cell line, MKN-45 cell-model.
5.1. Conclusions
Using an in-situ system, we showed that clinical isolates of H. pylori are able to form a biofilm on the human epithelial cell line, MKN-45. More detailed studies on differences of isolates with high and low abilities in biofilm formation may help understand the roles of bacterial factors in this process. Furthermore, we observed that H. pylori has the ability to bind to mucin, especially MUC5AC 5AC. So, presence of mucin in our biofilm formation assay, will better correspond to the natural conditions in host.
References
-
1.
Cammarota G, Sanguinetti M, Gallo A, Posteraro B. Review article: biofilm formation by Helicobacter pylori as a target for eradication of resistant infection. Aliment Pharmacol Ther. 2012;36(3):222-30. [PubMed ID: 22650647]. https://doi.org/10.1111/j.1365-2036.2012.05165.x.
-
2.
Dunne C, Dolan B, Clyne M. Factors that mediate colonization of the human stomach by Helicobacter pylori. World J Gastroenterol. 2014;20(19):5610-24. [PubMed ID: 24914320]. [PubMed Central ID: PMC4024769]. https://doi.org/10.3748/wjg.v20.i19.5610.
-
3.
Stark RM, Gerwig GJ, Pitman RS, Potts LF, Williams NA, Greenman J, et al. Biofilm formation by Helicobacter pylori. Let Appl Microbiol. 1999;28(2):121-6. https://doi.org/10.1046/j.1365-2672.1999.00481.x.
-
4.
Cellini L, Grande R, Traini T, Di Campli E, Di Bartolomeo S, Di Iorio D, et al. Biofilm formation and modulation of luxS and rpoD expression by Helicobacter pylori. Biofilms. 2005;2(2):119-27. https://doi.org/10.1017/S1479050505001845.
-
5.
Cole SP, Harwood J, Lee R, She R, Guiney DG. Characterization of monospecies biofilm formation by Helicobacter pylori. J Bacteriol. 2004;186(10):3124-32. [PubMed ID: 15126474]. [PubMed Central ID: PMC400600]. https://doi.org/10.1128/JB.186.10.3124-3132.2004.
-
6.
Di Campli E, Di Bartolomeo S, Grande R, Di Giulio M, Cellini L. Effects of extremely low-frequency electromagnetic fields on Helicobacter pylori biofilm. Curr Microbiol. 2010;60(6):412-8. [PubMed ID: 20033173]. https://doi.org/10.1007/s00284-009-9558-9.
-
7.
Grande R, Di Campli E, Di Bartolomeo S, Verginelli F, Di Giulio M, Baffoni M, et al. Helicobacter pylori biofilm: a protective environment for bacterial recombination. J Appl Microbiol. 2012;113(3):669-76. [PubMed ID: 22639839]. https://doi.org/10.1111/j.1365-2672.2012.05351.x.
-
8.
Yonezawa H, Osaki T, Hanawa T, Kurata S, Ochiai K, Kamiya S. Impact of Helicobacter pylori biofilm formation on clarithromycin susceptibility and generation of resistance mutations. PLoS One. 2013;8(9). e73301. [PubMed ID: 24039906]. [PubMed Central ID: PMC3765302]. https://doi.org/10.1371/journal.pone.0073301.
-
9.
Carron MA, Tran VR, Sugawa C, Coticchia JM. Identification of Helicobacter pylori biofilms in human gastric mucosa. J Gastrointest Surg. 2006;10(5):712-7. [PubMed ID: 16713544]. https://doi.org/10.1016/j.gassur.2005.10.019.
-
10.
Coticchia JM, Sugawa C, Tran VR, Gurrola J, Kowalski E, Carron MA. Presence and density of Helicobacter pylori biofilms in human gastric mucosa in patients with peptic ulcer disease. J Gastrointest Surg. 2006;10(6):883-9. [PubMed ID: 16769546]. https://doi.org/10.1016/j.gassur.2005.12.009.
-
11.
Lebeaux D, Chauhan A, Rendueles O, Beloin C. From in vitro to in vivo Models of Bacterial Biofilm-Related Infections. Pathogens. 2013;2(2):288-356. [PubMed ID: 25437038]. [PubMed Central ID: PMC4235718]. https://doi.org/10.3390/pathogens2020288.
-
12.
Roberts AE, Kragh KN, Bjarnsholt T, Diggle SP. The Limitations of In Vitro Experimentation in Understanding Biofilms and Chronic Infection. J Mol Biol. 2015;427(23):3646-61. [PubMed ID: 26344834]. https://doi.org/10.1016/j.jmb.2015.09.002.
-
13.
Salas-Jara MJ, Sanhueza EA, Retamal-Diaz A, Gonzalez C, Urrutia H, Garcia A. Probiotic Lactobacillus fermentum UCO-979C biofilm formation on AGS and Caco-2 cells and Helicobacter pylori inhibition. Biofouling. 2016;32(10):1245-57. [PubMed ID: 27834106]. https://doi.org/10.1080/08927014.2016.1249367.
-
14.
Ashrafi M, Novak-Frazer L, Bates M, Baguneid M, Alonso-Rasgado T, Xia G, et al. Validation of biofilm formation on human skin wound models and demonstration of clinically translatable bacteria-specific volatile signatures. Sci Rep. 2018;8(1):1-16. https://doi.org/10.1038/s41598-018-27504-z.
-
15.
Falsafi T, Favaedi R, Mahjoub F, Najafi M. Application of stool-PCR test for diagnosis of Helicobacter pylori infection in children. World J Gastroenterol. 2009;15(4):484-8. [PubMed ID: 19152455]. [PubMed Central ID: PMC2653372]. https://doi.org/10.3748/wjg.15.484.
-
16.
Attaran B, Falsafi T, Ghorbanmehr N. Effect of biofilm formation by clinical isolates of Helicobacter pylori on the efflux-mediated resistance to commonly used antibiotics. World J Gastroenterol. 2017;23(7):1163-70. [PubMed ID: 28275296]. [PubMed Central ID: PMC5323441]. https://doi.org/10.3748/wjg.v23.i7.1163.
-
17.
Grande R, Di Giulio M, Bessa LJ, Di Campli E, Baffoni M, Guarnieri S, et al. Extracellular DNA in Helicobacter pylori biofilm: a backstairs rumour. J Appl Microbiol. 2011;110(2):490-8. [PubMed ID: 21143715]. https://doi.org/10.1111/j.1365-2672.2010.04911.x.
-
18.
Dheepa M. Comparision of biofilm production and multiple drug resistance in clinical isolates of Acinetobacter baumanii from a tertiary care hospital in South India. Int J Pharm Biomed Sci. 2011;2(4):103-7.
-
19.
Attaran B, Falsafi T. Identification of Factors Associated with Biofilm Formation Ability in the Clinical Isolates of Helicobacter pylori. Iran J Biotechnol. 2017;15(1):58-66. [PubMed ID: 28959353]. [PubMed Central ID: PMC5582254]. https://doi.org/10.15171/ijb.1368.
-
20.
Falsafi T, Mahboubi M, Zargampour Z, Ahmadi M. Association between efficient adhesion to HEp-2 cells and severity of gastritis in Helicobacter pylori strains isolated from children. Iran J Microbiol. 2009;1(1):23-30.
-
21.
Hennequin C, Porcheray F, Waligora-Dupriet A, Collignon A, Barc M, Bourlioux P, et al. GroEL (Hsp60) of Clostridium difficile is involved in cell adherence. Microbiology (Reading). 2001;147(Pt 1):87-96. [PubMed ID: 11160803]. https://doi.org/10.1099/00221287-147-1-87.
-
22.
Falsafi T, Lavasani P, Basardeh I, Massarrat S, Landarani Z. Evaluation of an Iranian Home-made Helicobacter pylori Stool Antigen ELISA Kit. Jundishapur J Microbiol. 2014;7(6). e10629. [PubMed ID: 25371803]. [PubMed Central ID: PMC4217660]. https://doi.org/10.5812/jjm.10629.
-
23.
Rogers SL, Rogers GC. Culture of Drosophila S2 cells and their use for RNAi-mediated loss-of-function studies and immunofluorescence microscopy. Nat Protoc. 2008;3(4):606-11. [PubMed ID: 18388942]. https://doi.org/10.1038/nprot.2008.18.
-
24.
Kozakov D, Hall DR, Xia B, Porter KA, Padhorny D, Yueh C, et al. The ClusPro web server for protein-protein docking. Nat Protoc. 2017;12(2):255-78. [PubMed ID: 28079879]. [PubMed Central ID: PMC5540229]. https://doi.org/10.1038/nprot.2016.169.
-
25.
Desta IT, Porter KA, Xia B, Kozakov D, Vajda S. Performance and Its Limits in Rigid Body Protein-Protein Docking. Structure. 2020;28(9):1071-1081 e3. [PubMed ID: 32649857]. [PubMed Central ID: PMC7484347]. https://doi.org/10.1016/j.str.2020.06.006.
-
26.
Kozakov D, Beglov D, Bohnuud T, Mottarella SE, Xia B, Hall DR, et al. How good is automated protein docking? Proteins. 2013;81(12):2159-66. [PubMed ID: 23996272]. [PubMed Central ID: PMC3934018]. https://doi.org/10.1002/prot.24403.
-
27.
Vajda S, Yueh C, Beglov D, Bohnuud T, Mottarella SE, Xia B, et al. New additions to the ClusPro server motivated by CAPRI. Proteins. 2017;85(3):435-44. [PubMed ID: 27936493]. [PubMed Central ID: PMC5313348]. https://doi.org/10.1002/prot.25219.
-
28.
Meng EC, Pettersen EF, Couch GS, Huang CC, Ferrin TE. Tools for integrated sequence-structure analysis with UCSF Chimera. BMC Bioinformatics. 2006;7:339. [PubMed ID: 16836757]. [PubMed Central ID: PMC1570152]. https://doi.org/10.1186/1471-2105-7-339.
-
29.
Rizzato C, Torres J, Kasamatsu E, Camorlinga-Ponce M, Bravo MM, Canzian F, et al. Potential Role of Biofilm Formation in the Development of Digestive Tract Cancer With Special Reference to Helicobacter pylori Infection. Front Microbiol. 2019;10:846. [PubMed ID: 31110496]. [PubMed Central ID: PMC6501431]. https://doi.org/10.3389/fmicb.2019.00846.
-
30.
Schlaermann P, Toelle B, Berger H, Schmidt SC, Glanemann M, Ordemann J, et al. A novel human gastric primary cell culture system for modelling Helicobacter pylori infection in vitro. Gut. 2016;65(2):202-13. [PubMed ID: 25539675]. [PubMed Central ID: PMC4752654]. https://doi.org/10.1136/gutjnl-2014-307949.
-
31.
Cai J, Huang H, Song W, Hu H, Chen J, Zhang L, et al. Preparation and evaluation of lipid polymer nanoparticles for eradicating H. pylori biofilm and impairing antibacterial resistance in vitro. Int J Pharm. 2015;495(2):728-37. https://doi.org/10.1016/j.ijpharm.2015.09.055.
-
32.
Hathroubi S, Hu S, Ottemann KM. Genetic requirements and transcriptomics of Helicobacter pylori biofilm formation on abiotic and biotic surfaces. npj Biofilms Microbiomes. 2020;6(1). https://doi.org/10.1038/s41522-020-00167-3.
-
33.
Tan S, Tompkins LS, Amieva MR. Helicobacter pylori usurps cell polarity to turn the cell surface into a replicative niche. PLoS Pathog. 2009;5(5). e1000407. [PubMed ID: 19412339]. [PubMed Central ID: PMC2669173]. https://doi.org/10.1371/journal.ppat.1000407.
-
34.
Yonezawa H, Osaki T, Kurata S, Zaman C, Hanawa T, Kamiya S. Assessment of in vitro biofilm formation by Helicobacter pylori. J Gastroenterol Hepatol. 2010;25 Suppl 1:S90-4. [PubMed ID: 20586874]. https://doi.org/10.1111/j.1440-1746.2009.06213.x.
-
35.
McAuley JL, Linden SK, Png CW, King RM, Pennington HL, Gendler SJ, et al. MUC1 cell surface mucin is a critical element of the mucosal barrier to infection. J Clin Invest. 2007;117(8):2313-24. [PubMed ID: 17641781]. [PubMed Central ID: PMC1913485]. https://doi.org/10.1172/JCI26705.
-
36.
Perrais M, Rousseaux C, Ducourouble MP, Courcol R, Vincent P, Jonckheere N, et al. Helicobacter pylori urease and flagellin alter mucin gene expression in human gastric cancer cells. Gastric Cancer. 2014;17(2):235-46. [PubMed ID: 23703470]. https://doi.org/10.1007/s10120-013-0267-5.
-
37.
Linden S, Nordman H, Hedenbro J, Hurtig M, Boren T, Carlstedt I. Strain- and blood group-dependent binding of Helicobacter pylori to human gastric MUC5AC glycoforms. Gastroenterology. 2002;123(6):1923-30. [PubMed ID: 12454849]. https://doi.org/10.1053/gast.2002.37076.
-
38.
Guang W, Czinn SJ, Blanchard TG, Kim KC, Lillehoj EP. Genetic regulation of MUC1 expression by Helicobacter pylori in gastric cancer cells. Biochem Biophys Res Commun. 2014;445(1):145-50. [PubMed ID: 24491543]. [PubMed Central ID: PMC3964589]. https://doi.org/10.1016/j.bbrc.2014.01.142.
-
39.
Niv Y. Helicobacter pylori and gastric mucin expression: A systematic review and meta-analysis. World J Gastroenterol. 2015;21(31):9430-6. [PubMed ID: 26309370]. [PubMed Central ID: PMC4541396]. https://doi.org/10.3748/wjg.v21.i31.9430.
-
40.
Linden SK, Wickstrom C, Lindell G, Gilshenan K, Carlstedt I. Four modes of adhesion are used during Helicobacter pylori binding to human mucins in the oral and gastric niches. Helicobacter. 2008;13(2):81-93. [PubMed ID: 18321298]. https://doi.org/10.1111/j.1523-5378.2008.00587.x.
-
41.
Matsuda K, Yamauchi K, Matsumoto T, Sano K, Yamaoka Y, Ota H. Quantitative analysis of the effect of Helicobacter pylori on the expressions of SOX2, CDX2, MUC2, MUC5AC, MUC6, TFF1, TFF2, and TFF3 mRNAs in human gastric carcinoma cells. Scand J Gastroenterol. 2008;43(1):25-33. [PubMed ID: 18938748]. [PubMed Central ID: PMC3128253]. https://doi.org/10.1080/00365520701579795.
-
42.
Linden SK, Driessen KM, McGuckin MA. Improved in vitro model systems for gastrointestinal infection by choice of cell line, pH, microaerobic conditions, and optimization of culture conditions. Helicobacter. 2007;12(4):341-53. [PubMed ID: 17669108]. https://doi.org/10.1111/j.1523-5378.2007.00509.x.