Abstract
Background:
microRNAs (miRNAs) have been highlighted as potential circulating biomarkers and therapeutic targets for breast cancer (BC). miRNAs have emerged as an extremely promising new class of biomarkers. They have been demonstrated to be remarkably stable in human blood. miR-155 has more than 4 hundred predicted gene targets. The miR-155 plays an essential role in the pathogenesis of breast cancer. The aim of this study was to assess the diagnostic value of miR-155 expression in the serum of patients with breast cancer according to the molecular subtypes of BC.Methods:
90 samples were classified into different groups with respect to their immunohistochemical (IHC) characteristics: estrogen receptor (ER) positive and/or progesterone receptor (PR) positive group (Luminal A), human epidermal growth factor receptor2 (HER2) positive group (as Luminal B), and Triple negative group (TNBC). The complementary DNA (cDNA) was synthesized in line with the guidelines of the Kit Company. A real-time PCR method was performed as the expression assay.Results:
A substantial difference was observed between the phrase standard of miR-155 in patients with the molecular subtypes of breast cancer. The subtypes of Luminal A, Luminal B, and TNBC showed respectively 6 fold, 7 fold, and 14 fold increased expression (P < 0.001). Receiver operating characteristic (ROC) analysis revealed that miR-155 had a considerable diagnostic accuracy for Luminal A with area under the curve (AUC) of 0.875 (sensitivity 100%, specificity 68.63%), for Luminal B with AUC of 0.886 (sensitivity 100%, specificity 62.75%), and for TNBC with AUC of 0.982 (sensitivity 100%, specificity 92.16%)Conclusions:
The study found that increased mir-155 expression was significantly associated with BC. The study results showed the mir-155 expression has extremely high sensitivity and specificity for the diagnosis of subtypes of breast tumor. Therefore, checking the serum levels of miR-155 expression in patients with breast cancer may be helpful. Yet, more studies are required to investigate this relationship.Keywords
1. Background
Breast cancer (BC) is the most prevalent cancer in women comprising about 16% of all cancer cases in women (1, 2). A subtype of BC, called triple negative, without ER/PR/Her2 expression grows without the need for external growth hormones (3), which makes tumors grow quickly, Luminal A: ER and/or PR positive and HER2-negative and Luminal B: ER and/or PR and HER2 positive (4). These markers are usually proteins but they can also be other detectable molecules, such as oligonucleotides (5).
miRNAs are short 20 to 25 nucleotide, non-coding RNA strands that bind to messenger RNAs (mRNAs) to inhibit translation through post-transcriptional modifications and induced mRNA degradation (6, 7). The emerging evidence indicating that certain microRNAs (miRNAs) are extensively involved in BC progression has led to the search for good miRNA candidates for classifying tumors (8). Recently, it has been found that these miRNAs in serum proteins, lipoproteins, and proteins make connections called Argonaute-2 causing the miRNAs in serum safeguarded and protected from ribonucleases (9). In addition, close-by markets and protection by secreting substances such as objects are imported Apoptotic and exosomes in the blood serum and secretory vesicles containing miRNAs addition; it can act as carriers of messages between cells miR-155, a miRNA to be involved in lymphoma is also rising to possess a role in the progression of solid cancer (10).
The miR-155 was first described in the progression of lymphoma (11). miR-155 has more than 4 hundred predicted gene targets. Generally, there is now a growing function of miR-155 in BC (12). miR-155 is one of the most multifunctional miRNAs whose overexpression has been found to be related to different types of cancers, including BC (13). A study has demonstrated the association of elevated miR-155 with late stage and poor overall survival in a few types of malignancy (14). As a miRNA, miR-155 is an oncogenic property, just about all signs of cancers in the breast malignancy affects different routes (12, 15).
2. Objectives
The current study aimed to examine the diagnostic value of mir155 expression in the serum of patients with BC, according to molecular subtypes of breasts cancer, including (Luminal A/B, overexpression of Her2, and their triple-negative type), and compare them with a control sex-age-gender matched group.
3. Methods
3.1. Study Population
In a case- control study, 90 female patients with BC who had been diagnosed by pathological examinations in Ahvaz from 2012 to 2015 were recruited. 70 healthy subjects with similar age who received medical tests were selected as controls.
3.2. Ethics
The research protocol was approved by a suitably constituted Ethics Committee of Ahvaz Jundishapur University of Medical Sciences.
3.3. Sample Preparation
5 cc blood samples were taken from the healthy individual SST tube (for serum). The samples were centrifuged at 2500 rpm for 5 minutes at room temperature. Cruor was then transferred to a brand new tube and centrifuged at 3000 rpm for 5 minutes at room temperature. The samples were aliquoted into Microcentrifuge tubes and then kept at -80°C.
3.4. miRNA Isolation
miRNAs from serum samples were extracted using miRNeasy Serum/Plasma Kit (Qiagen, Germany). Extraction was performed in accordance with the manufacturer’s instructions at -80°C.
3.5. Reverse Transcription (RT) and Quantitative PCR
Using a High Capacity cDNA kit (Life Technologies, America) for miRNA isolated from peripheral blood serum, cDNA was prepared to continue the next step in product temperature at -20°C. Proliferative responses were conducted in a final volume of 20 µL, and a double in 96-well plates and devices using Real-Time PCR (Steponeplus ABI, America) were performed. For Mixed reaction, 10 µL SYBR- Green PCR (Master Mix Takara), 1 mL primer (Forward), and reverse (Reverse) specific for each gene, 5 µL of cDNA were mixed and finally by adding distilled water reached the final volume of 20 µL. The primers for miR-155 were used as follows: forward: 5′-GATACTCATAAGGCACGCGG-3′ and reverse: 5′-GTGCAGGGTCCGAGGT-3′. To compare expression levels of miR-155 in the serum of patients with BC, the relative amount of miR-155 was normalized against SNORD 47. In the study, we calculated the fold-change between breast cancer patients and normal controls for miR-155 by the 2ΔΔCt method.
3.6. Statistical Analysis
Laboratory data were analyzed by SPSS®ver 20.0 for Windows®. In the descriptive study, data were expressed as percentages and analyzed by analysis of variance (ANOVA). Receiver operating characteristic (ROC) curve analysis was used to assess the diagnostic accuracy. The area under the ROC curve (AUC) was then estimated. The P < 0.05 was considered statistically significant.
4. Results
Among 90 female patients, Luminal A comprised 23 (25.55%) cases, Luminal B 50 (55.55%) cases, and triple negative 17 (18.88%) cases. 70 cases of healthy subjects were selected as controls. The average age of BC patients was 46 years (SD = 13.30), and the average age of healthy subjects was 46.65 years (SD = 12.39).
We analyzed the relative expression of miR-155 in BC patients with ER positive and/or PR positive group (as Luminal A) and observed high expression of miR-155 (about 6 fold) (P < 0.001). In Luminal B tumors that tended to be ER-positive and/or PR-positive and HER2-positive (as group II), miR-155 was also over-expressed (about 7 fold) (P < 0.001). Triple negative group (TNBC) that lacked ER/PR/Her2 (as group III) also showed miR-155 over-expression (about 14 fold) that was statistically significant (P < 0.001) (Table 1).
miR-155 Expressions in Patients with Breast Cancer, According to Molecular Subtypes of Breast Cancer
| Subtypes of Breast Cancer | No. (%) | miR-155 Expression (mean ± SD) | Std. Error | 95% Confidence Interval |
|---|---|---|---|---|
| Luminal A | 23 | 22.59 ± 12.72 | 2.78 | 17.06 to 28.11 |
| Luminal B | 50 | 25.77 ± 14.44 | 1.88 | 22.02 to 29.52 |
| Triple negative | 17 | 40.61 ± 10.30 | 3.23 | 34.18 to 47.04 |
To further evaluate the diagnostic proficiency of miR-155 for Luminal A breast cancer, the ROC curve was plotted, which can be seen in Figure 1. The AUC and confidence interval (CI) for miR-155 were as follows: 0.875 (0.77 - 0.94) with 100% sensitivity and 68.63% % specificity indicating high sensitivity and specificity of miR-155 for Luminal A breast cancer diagnosis.
ROC Curve to Evaluate the Application Value of miR-155 for Luminal A Breast Cancer Diagnosis
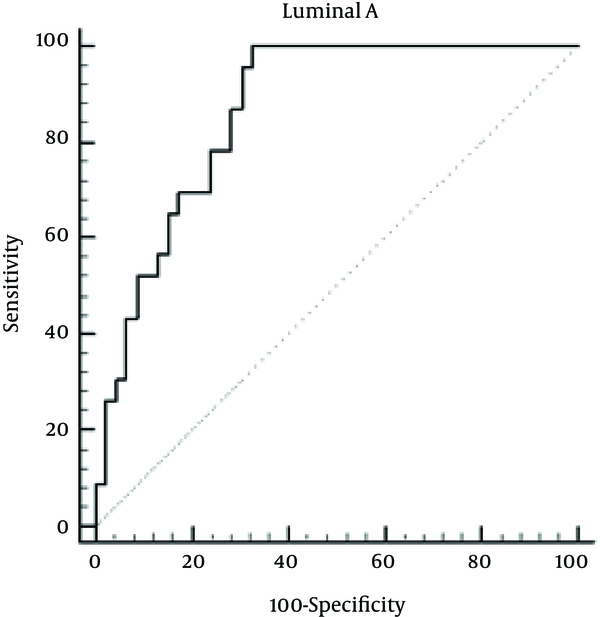
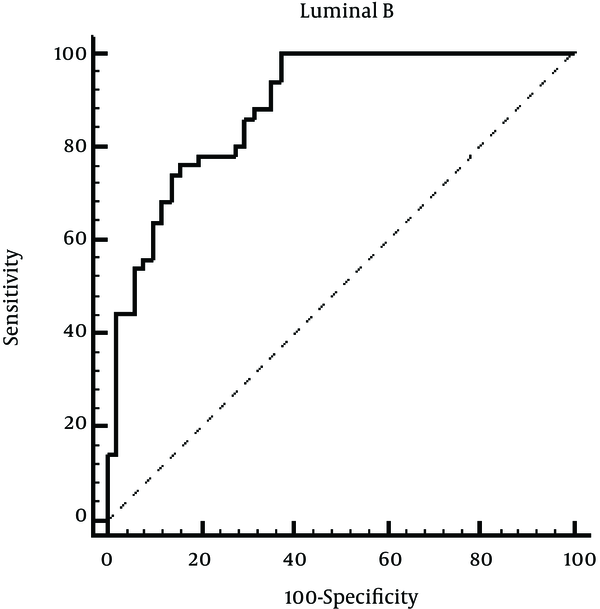
For investigating the diagnostic proficiency of miR-155 for Luminal B breast cancer, the ROC curve was plotted, which can be seen in Figure 2. The AUC and confidence interval (CI) for miR-155 were as follows: 0.886 (0.807 - 0.940) with 100% sensitivity and 62.75% specificity indicating high sensitivity and specificity of miR-155 for Luminal B breast cancer diagnosis.
ROC Curve to Evaluate the Application Value of miR-155 for Luminal B Diagnosis
For investigating the diagnostic proficiency of miR-155 for Triple negative, The ROC curve was plotted, which can be seen in Figure 3. The AUC and confidence interval (CI) for miR-155 were as follows: 0.982 (0.915 - 0.999) with 100% sensitivity and 92.16% specificity indicating high sensitivity and specificity of miR-155 for Triple negative breast cancer diagnosis.
ROC Curve to Evaluate the Application Value of miR-155 for Triple Negative Breast Cancer Diagnosis

5. Discussion
In recent years, several studies have reported that the levels of circulating miRNA can be used to differentiate healthy from malignancy patient blood samples (16). miRNAs have been highlighted as potential circulating biomarkers and therapeutic targets for BC (17). Other markers such as ER, PR, and Her2/neu position would indicate ideal adjunct remedy situation (9). Several studies linked miR-155 to inflammation in cancer (18). miR-155 expression is significantly increased in many cancers, including breast cancer (17).
The current study aimed to assess the diagnostic value of mir-155 expression in the serum of patients with breast cancer according to the molecular subtypes of BC. The obtained results showed that serum miR-155 was over-expressed in three subtypes of malignant breast cancer, which indicated that serum miR-155 might be taken as prognosis biomarkers for BC with high sensitivity and specificity for the diagnosis of subtypes of BC. miR-155 transgenic mice develop B-cell lymphoma, while miR-155 knockout mice exhibit damaged immune function (19). The majority of validated miR-155 functional biology and protein targets define the value of miR-155 in immunology and various kinds of lymphoma and BC (20). It is reported that miR-155 affects SOCS1 and STAT3 in BC and therefore activates the inflammatory cascade than the relationship between miR-155, inflammation, and breast cancer (21). In many classes of human breast malignant cells, the expression group miR-155 as well as factors of infection, tumor, such as interleukin-6 or IFN-γ- induced, in the course of miR-155 inhibit SOCS1, and stimulate the activity of the JAK2/STAT3 and promote inflammation and expansion of more tumors (22). A study conducted by Baccim et al. showed that miR-155 was associated with subtype of BC (23). Roth et al. (24), Wang et al. (25) and Sun et al. (26) showed high level of miR-155 was associated with breast cancer. The results showed significant differences in mir-155 expressions between patients with various subtypes of breast cancers. The current experiment was consistent with the study of Baccim et al. owing to a lack of diagnostic value of circulating miR-155 in patients with subtype of BC; we further performed ROC curve analysis that showed mir155 had very high sensitivity and high specificity for the diagnosis of subtypes of BC.
In the current study, there was a difference between the serum expression levels of miR-155 in patients with breast cancer based on Molecular Subtypes of Breast Cancer, indicating high sensitivity and specificity of miR-155 for the diagnosis of subtypes of BC.
5.1. Conclusions
In this study, according to molecular subtypes of breasts cancer and ROC curve analysis, it was disclosed that serum miR-155 expression has moderate accuracy and reliability in early analysis of breast cancer. mir-155 expression has very high sensitivity for the diagnosis of subtypes of breast cancer, but it has a different specificity in other subtypes of breast cancer. The study results showed that checking the serum levels of miR-155 expression in patients with breast cancer might be helpful. Expression amount of some miRNAs not only in tissues but also in circulation such as in plasma can accurately echo tumor malignancy. It is therefore recommended that similar studies be conducted in other populations with large number of cases.
Acknowledgements
References
-
1.
Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61(2):69-90. [PubMed ID: 21296855]. https://doi.org/10.3322/caac.20107.
-
2.
Hamilton AM, Aidoudi-Ahmed S, Sharma S, Kotamraju VR, Foster PJ, Sugahara KN, et al. Nanoparticles coated with the tumor-penetrating peptide iRGD reduce experimental breast cancer metastasis in the brain. J Mol Med (Berl). 2015;93(9):991-1001. [PubMed ID: 25869026]. https://doi.org/10.1007/s00109-015-1279-x.
-
3.
Metzger-Filho O, Sun Z, Viale G, Price KN, Crivellari D, Snyder RD, et al. Patterns of Recurrence and outcome according to breast cancer subtypes in lymph node-negative disease: results from international breast cancer study group trials VIII and IX. J Clin Oncol. 2013;31(25):3083-90. [PubMed ID: 23897954]. https://doi.org/10.1200/JCO.2012.46.1574.
-
4.
Cancer Genome Atlas N. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490(7418):61-70. [PubMed ID: 23000897]. https://doi.org/10.1038/nature11412.
-
5.
Yang XR, Chang-Claude J, Goode EL, Couch FJ, Nevanlinna H, Milne RL, et al. Associations of breast cancer risk factors with tumor subtypes: a pooled analysis from the Breast Cancer Association Consortium studies. J Natl Cancer Inst. 2011;103(3):250-63. [PubMed ID: 21191117]. https://doi.org/10.1093/jnci/djq526.
-
6.
Ghildiyal M, Zamore PD. Small silencing RNAs: an expanding universe. Nat Rev Genet. 2009;10(2):94-108. [PubMed ID: 19148191]. https://doi.org/10.1038/nrg2504.
-
7.
Tan G, Tang X, Tang F. The role of microRNAs in nasopharyngeal carcinoma. Tumour Biol. 2015;36(1):69-79. [PubMed ID: 25427638]. https://doi.org/10.1007/s13277-014-2847-3.
-
8.
McManus MT. MicroRNAs and cancer. Semin Cancer Biol. 2003;13(4):253-8. [PubMed ID: 14563119].
-
9.
Ortiz-Quintero B. Cell-free microRNAs in blood and other body fluids, as cancer biomarkers. Cell Prolif. 2016;49(3):281-303. [PubMed ID: 27218664]. https://doi.org/10.1111/cpr.12262.
-
10.
Kluiver J, Poppema S, de Jong D, Blokzijl T, Harms G, Jacobs S, et al. BIC and miR-155 are highly expressed in Hodgkin, primary mediastinal and diffuse large B cell lymphomas. J Pathol. 2005;207(2):243-9. [PubMed ID: 16041695]. https://doi.org/10.1002/path.1825.
-
11.
Clurman BE, Hayward WS. Multiple proto-oncogene activations in avian leukosis virus-induced lymphomas: evidence for stage-specific events. Mol Cell Biol. 1989;9(6):2657-64. https://doi.org/10.1128/mcb.9.6.2657.
-
12.
Chen J, Wang BC, Tang JH. Clinical significance of microRNA-155 expression in human breast cancer. J Surg Oncol. 2012;106(3):260-6. [PubMed ID: 22105810]. https://doi.org/10.1002/jso.22153.
-
13.
Kong W, Yang H, He L, Zhao JJ, Coppola D, Dalton WS, et al. MicroRNA-155 is regulated by the transforming growth factor beta/Smad pathway and contributes to epithelial cell plasticity by targeting RhoA. Mol Cell Biol. 2008;28(22):6773-84. [PubMed ID: 18794355]. https://doi.org/10.1128/MCB.00941-08.
-
14.
Xiang X, Zhuang X, Ju S, Zhang S, Jiang H, Mu J, et al. miR-155 promotes macroscopic tumor formation yet inhibits tumor dissemination from mammary fat pads to the lung by preventing EMT. Oncogene. 2011;30(31):3440-53. [PubMed ID: 21460854]. https://doi.org/10.1038/onc.2011.54.
-
15.
Mattiske S, Suetani RJ, Neilsen PM, Callen DF. The oncogenic role of miR-155 in breast cancer. Cancer Epidemiol Biomarkers Prev. 2012;21(8):1236-43. [PubMed ID: 22736789]. https://doi.org/10.1158/1055-9965.EPI-12-0173.
-
16.
Yu DC, Li QG, Ding XW, Ding YT. Circulating microRNAs: potential biomarkers for cancer. Int J Mol Sci. 2011;12(3):2055-63. [PubMed ID: 21673939]. https://doi.org/10.3390/ijms12032055.
-
17.
Farazi TA, Horlings HM, Ten Hoeve JJ, Mihailovic A, Halfwerk H, Morozov P, et al. MicroRNA sequence and expression analysis in breast tumors by deep sequencing. Cancer Res. 2011;71(13):4443-53. [PubMed ID: 21586611]. https://doi.org/10.1158/0008-5472.CAN-11-0608.
-
18.
Azizian A, Gruber J, Ghadimi BM, Gaedcke J. MicroRNA in rectal cancer. World J Gastrointest Oncol. 2016;8(5):416-26. [PubMed ID: 27190581]. https://doi.org/10.4251/wjgo.v8.i5.416.
-
19.
Slezak-Prochazka I, Kluiver J, de Jong D, Smigielska-Czepiel K, Kortman G, Winkle M, et al. Inhibition of the miR-155 target NIAM phenocopies the growth promoting effect of miR-155 in B-cell lymphoma. Oncotarget. 2016;7(3):2391-400. [PubMed ID: 26497687]. https://doi.org/10.18632/oncotarget.6165.
-
20.
Hemmatzadeh M, Mohammadi H, Jadidi-Niaragh F, Asghari F, Yousefi M. The role of oncomirs in the pathogenesis and treatment of breast cancer. Biomed Pharmacother. 2016;78:129-39. [PubMed ID: 26898434]. https://doi.org/10.1016/j.biopha.2016.01.026.
-
21.
Jiang S, Zhang HW, Lu MH, He XH, Li Y, Gu H, et al. MicroRNA-155 functions as an OncomiR in breast cancer by targeting the suppressor of cytokine signaling 1 gene. Cancer Res. 2010;70(8):3119-27. [PubMed ID: 20354188]. https://doi.org/10.1158/0008-5472.CAN-09-4250.
-
22.
Persson JL. miR-155 meets the JAK/STAT pathway. Cell Cycle. 2013;12(14):2170. [PubMed ID: 23803728]. https://doi.org/10.4161/cc.25548.
-
23.
Bacci M, Giannoni E, Fearns A, Ribas R, Gao Q, Taddei ML, et al. miR-155 Drives Metabolic Reprogramming of ER+ Breast Cancer Cells Following Long-Term Estrogen Deprivation and Predicts Clinical Response to Aromatase Inhibitors. Cancer Res. 2016;76(6):1615-26. [PubMed ID: 26795347]. https://doi.org/10.1158/0008-5472.CAN-15-2038.
-
24.
Roth C, Rack B, Muller V, Janni W, Pantel K, Schwarzenbach H. Circulating microRNAs as blood-based markers for patients with primary and metastatic breast cancer. Breast Cancer Res. 2010;12(6):R90. [PubMed ID: 21047409]. https://doi.org/10.1186/bcr2766.
-
25.
Wang F, Zheng Z, Guo J, Ding X. Correlation and quantitation of microRNA aberrant expression in tissues and sera from patients with breast tumor. Gynecol Oncol. 2010;119(3):586-93. [PubMed ID: 20801493]. https://doi.org/10.1016/j.ygyno.2010.07.021.
-
26.
Sun Y, Wang M, Lin G, Sun S, Li X, Qi J, et al. Serum microRNA-155 as a potential biomarker to track disease in breast cancer. PLoS One. 2012;7(10). e47003. [PubMed ID: 23071695]. https://doi.org/10.1371/journal.pone.0047003.