Abstract
Background:
Paint coating systems are widely implemented on different surfaces for both aspects of decoration and protection against corrosion. Due to the presence of organic compounds, the growth of microorganisms is more likely to take place in paints, such as automotive paint. In the process of automotive painting, 20% - 60% of the paint does not expose to the automotive body, which is washed using water and would lead to the painting sludge formation. Paint sludge is considered one of the hazardous wastes from the automotive industry, which is finally landfilled or incinerated.Objectives:
Despite the presence of inhibiting compounds in paint sludge, such as heavy metals and biocides, the objective of this study was to isolate and identify microorganisms in the sludge culture.Methods:
The microorganisms were isolated using serial dilutions, direct cultivation, and enrichment methods in basic salt cultivation media. Then, their biochemical and molecular specifications were investigated.Results:
The number of microorganisms counted in paint sludge was approximately around 1 × 104 CFU/mL, and six isolated colonies were finally obtained.Conclusions:
The main isolated microbial consortium from paint sludge included Pseudomonas aeruginosa, Staphylococcus haemolyticus, Micrococcus yunnanensis, Rothia amarae, Gordonia terrae, and Brevibacillus agri. Nearly 83% of the isolated strains were Gram-positive.Keywords
Paint Sludge Automotive Industry Cultivation Media Six Bacterial Colonies Resistant Microorganisms
1. Background
Paint is a synthetic material used as a substrate in the texture of furniture and other things (1). Paint is applied using a brush, roller, or spray as a thin layer on wood, metals, and stones to protect against corrosion (2) and create excellent decoration effect. The vehicle paint is a mixture of binders, pigments, solvents, and additives (3). Binders cause adhesion in paint particles, pigments create color and prevent corrosion, solvents lead to the dispersion of paint, and additives strengthen brushing and resistance properties (4). Automotive paint consists of a multilayer coating: primer as an anti-corrosion layer, a base coating that paints the automotive body, and a clear coating for a radiant appearance and UV protection (5). To produce the final automotive color, two types of paints are required: water-based and solvent-based (6). Methods for painting the automotive body include overspray, immersion, and powder methods (7). In overspray staining, only 50% - 80% of the paint reaches the automotive body (6), and 20% - 60% of the paint is transmitted as surplus (8) to the room with airflow and is successively removed using rotary washing water. The detergents added to the wash water separate the flocculants and coagulants from excess paint and contribute to the separation of paint from water (6). The mixture of water and spray paint, called paint sludge, is collected in a sludge pit (7).
Today, with the increasing number of vehicles around the world, the production of paint sludge is growing (6). Each factory produces about 10 - 15 tones sludge per day, containing some elements and contaminants (9). The combination of volatile organic compounds in paints can cause environmental pollution in the short or long-term (10). Ingredients such as antifouling Tributyltin (TBT) in the paint are highly toxic to aquatic environments (2). Paint sludge is also a serious hazard to the health and environment, and in most cases, its production is inevitable (8). Eighty percent of environmental concerns in automotive painting factories are related to volatile organic compounds (VOCs), carbon dioxide (CO2), and heavy metals. As a waste, paint sludge contains heavy metals and dissoluble organic carbon (DOC) of over 3,700 mg/L and, thus, is extremely hazardous. Paint sludge is classified in the EU code for waste disposal 080113 and is considered a waste with hazardous characteristics (6).
Salihoglu et al. investigated the possibility of composting automotive water-based paint sludge with sewage sludge of the same industry as substrate and corncob as a bulking agent in sex reactors.
Their results showed that carbon and nitrogen, organic matter and BTEX, nickel and tin, the ratio decreased (11). Ghomi Avili et al. (12) investigated chromium removal from automotive paint sludge using biological sludge and Eisenia fetida worms. The results showed that the Cr+6 concentration fell from 6 mg/kg to less than 0.2 mg/kg, pH decreased from 7.8 to 7.3, volatile solids decreased from 80.4% to 37%, and the C/N ratio decreased from about 27 to 14.3 after 90 days.
So far, various isolator microbial species have been isolated and identified from paint (13). Some organic and inorganic compounds are toxic to microorganisms. High quantities of heavy metals (14) and biocides in the paint can inhibit the metabolic activity of bacteria and fungi. Biocides can affect the growth of microorganisms through degradation of the cell membrane, inhibition of metabolic reactions, alteration of intracellular pH, and accumulation of toxic anions (15). Despite the presence of inhibitors in automotive paint sludge, the possibility of the isolation and identification of resistant microorganisms in the sludge was investigated in this study.
2. Objectives
Lots of research have been conducted on the isolation and identification of microorganisms from petroleum sludge, sewage sludge, contaminated coastal soils, mine soils, contaminated sediments, municipal waste, and wastewater sites for use in bioremediation. In the current study, the possibility of isolation and identification of resistant microorganisms in paint sludge was investigated.
3. Methods
3.1. Sampling
Paint sludge samples were collected from the site of an automotive painting factory under aseptic conditions using sterile spatula gathered inside 100 mL sterile Falcon tubes. The samples were then transferred to the laboratory at a temperature of 4°C.
3.2. Methods for Isolation of Microorganisms
3.2.1. Isolation by the Enrichment Method
Conventional salt-based media such as Bushnell Haas medium (BHMS) (16), mineral salt medium (MSM) (17), 9KFe2+ (18), 9Ks, and 9KNa2S2O3 (19) were used to isolate a wide range of bacteria. Culture media were prepared according to Table 1. Subsequently, 2% (w/v) of sterile paint sludge was added as a single source of carbon and energy. With some modifications in the amount of inoculation, 15 g of a paint sludge sample under aseptic conditions was added to the medium and incubated at 30 ± 2°C in a shaker incubator at 110 rpm for five consecutive days. To create colonies, 10 - 15 g of agar and 2% (w/v) sterile paint sludge were added to produce a solid medium. Then, 0.1 mL of each salt medium of Erlenmeyer cultured (triplicate of each medium) in nutrient agar (NA), potato-dextrose agar (PDA), and plate count agar (PCA) using the pour plate and steric methods and incubated at 30 ± 2°C for five days (20, 21).
Composition of Salt Media for Isolation of Microorganisms
| Values | |
|---|---|
| BHMS media | |
| MgSO4.H2O, g | 0.2 |
| CaCl2.H2O, g | 0.002 |
| KH2PO4, g | 1 |
| K2HPO4, g | 1 |
| NH4NO3, g | 1 |
| FeCl3, g | 0.05 |
| Distilled water, cc | 1000 |
| pH | 7 |
| 9KFe2+ media | |
| (NH4)2SO4, g | 3 |
| KCL, g | 0.1 |
| K2HPO4, g | 0.5 |
| MgSO4.7H2O, g | 0.5 |
| Ca(NO3)2, g | 0.01 |
| Distilled water, cc | 700 |
| H2SO4 (w/v), N | 10 |
| FeSO4.7H2O, g | 44.22 |
| Distilled water, cc | 300 |
| pH | 1 - 2 |
| MSM media | |
| K2HPO4, g | 0.05 |
| MgSO4.H2O, g | 0.5 |
| Ca(NO3)2, g | 0.01 |
| KCl, g | 0.1 |
| Na2SO4.10H2O, g | 3. |
| (NH4)2SO4 | 3 |
| Distilled water, cc | 1000 |
| pH | 7 ± 0.2 |
| 9KS media | |
| (NH4)2SO4, g | 3 |
| KCl, g | 0.1 |
| K2HPO4, g | 0.5 |
| MgSO4.7H2O, g | 0.5 |
| Ca(NO3)2, g | 0.01 |
| Sulfur or thiosulfate, g | 10 |
| Distilled water, cc | 1000 |
| H2SO4, M | 2 |
| pH | 6.5 |
3.2.2. Isolation by Direct Cultivation and Serial Dilution Method
Ten g of paint sludge sample was poured into a 250 cc Erlenmeyer under aseptic conditions, and 90 mL of sterile distilled water was added. The Erlenmeyer was covered with sterile cotton and homogenized with a shaker. Then, a portion of 1 mL was added to a tube containing 9 mL of sterile distilled water along with shaking. It continued serially to a dilution of 10 - 5. Then, 1 mL of each dilution and direct sample were cultured (triplicate of each dilution) on a plate containing NA and PDA. The plates were incubated at 30 ± 2°C for five days (22).
3.3. Morphological Properties
Gram-positive and Gram-negative strains were detected by Gram staining and microscopic observation.
3.4. Biochemical Properties
Species were isolated according to Bergey’s manual of determinative bacteriology (23).
3.5. Molecular Identification
The 16SrRNA gene sequencing was performed by genomic extraction using Favorgen’s Mini Kit Genetic DNA extraction kit. A polymerase chain reaction (PCR) was done to determine 16SrRNA gene proliferation using primers 4F: 5’-TATCGGAGAGTTTGATCCTGG-3’ and 1541r: 5’-AAGGAGGGATCCAGCCGCA-3. The PCR program was performed in 35 cycles (24), as given in Table 2. The PCR products were electrophoresed, and after observing the proper band, they were purified by Favorgen’s Purification Mini kit. The DNA concentration after purification was measured at 260 nm. The purified PCR products were sent by the Iranian Research Organization for Science and Technology to Bioneer Company in Korea for sequencing. The sequencing was performed using primers as follows: 16r339: 5’-ACTGCTGCCTCCCGTAGGAG-3’, 27f: 5’-GAGTTTGATCCTGGCTCAG-3’, 704f: 5’-GTAGCGGTGAAATGCGTAGA-3’and 16f358: 5’-CTCCTACGGGAGGCAGCAG-3’. The obtained sequences were compared with nucleotide sequences available in valid databases, including NCBI and Eztaxon. Then, the phylogenetic trees were plotted using Mega version 6 software (25, 26).
Polymerase Chain Reaction Program Protocol
| Process | Temperature, °C | Time |
|---|---|---|
| Initial denaturation | 95 | 3’ |
| Denaturation | 93 | 45” |
| Annealing | 58 | 60” |
| Extension | 72 | 90” |
4. Results
The number of microorganisms counted with the plate count agar method was 1 × 104 CFU/mL. Six colonies were isolated on a saline culture medium, including BHMS, MSM, and 9KNa2S2O3, and nutrient agar medium using direct and serial methods., using the direct and serial methods. For Gram staining, an expansion of isolated colonies was prepared, and the isolation of species was performed based on Bergey’s manual. Some morphological and biochemical properties of the isolated microorganisms are given in Table 3.
Morphological and Biochemical Properties of the Isolated Microorganisms
| ID | aA | bB | cC | dD | eE | fF |
|---|---|---|---|---|---|---|
| Medium | BHMS | 9K | MSM | NB serial | NB serial | NB direct |
| Gram stain | Gram- | Gram+ | Gram+ | Gram+ | Gram+ | Gram+ |
| Morphology | Bacilli | Cocci | Cocci | Spherical | Coccoid | Rods |
| Colony shape | Circular | Circular | Circular | Circular | Rough | Flat |
| Colony color | Gray | White | Yellow | Cream | Orange | Gray |
| Size, mm | 2 - 2.5 | 0.5 - 1 | 0.1.5 - 3 | 1 | 2 - 2.5 | 2 - 4 |
| Catalase | + | + | + | + | + | + |
| Oxidase | - | - | - | - | - | + |
| Coagulase | + | - | ND | ND | ND | ND |
| OF | Oxidative | Ferment | Inert | Ferment | ND | + |
| Motility | + | ND | ND | ND | ND | + |
| Indole | - | ND | ND | ND | ND | - |
| Urease | ND | + | - | - | + | - |
| Hemolysis | ND | ND | ND | ND | ND | Alfa |
| Nitrate reduce | ND | + | + | - | + | - |
| Starch | ND | ND | ND | ND | - | + |
| Lipase | + | ND | - | - | + | ND |
| Gelatin hydrolysis | + | ND | ND | ND | ND | ND |
| Denitrification | + | ND | ND | ND | ND | ND |
| Hemolysis | ND | + | ND | ND | ND | ND |
| ONPG | ND | - | ND | ND | ND | ND |
| MR/VP | ND | ND | -/- | ND | ND | ND/- |
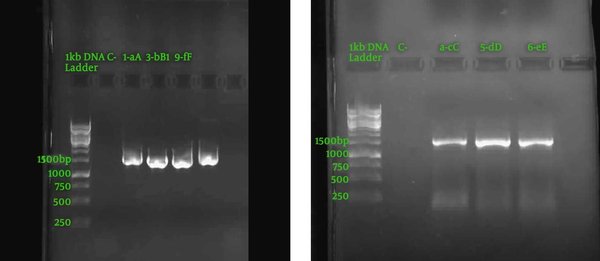
To investigate the PCR products, electrophoresis was performed, and 1,500 nucleotide fragments were observed, according to Figure 1. The DNA concentration after purification at 260 nm was measured, and the results are presented in Table 4. The analysis disclosed sequences with different nucleotides. These sequences were compared by basic local alignment search tool (BLAST) with the nucleotide sequences available in valid databases, such as the National Center for Biotechnology Information (NCBI), and the results of the similarity percentage of the strains with the existing ones are presented in Table 5. Figure 2 indicates an example of a phylogenetic tree using Mega version 6 software.
The Concentration of Purified DNA
| Strain Code | DNA Concentration, ng/µL |
|---|---|
| Strain aA | 154 |
| Strain bB | 87.5 |
| Strain cC | 89.6 |
| Strain dD | 70.7 |
| Strain eE | 93.8 |
PCR product electrophoresis
The phylogenetic tree of the fF-Brevibacillus strain plotted with Mega software version 6.

The Comparison of the Similarity Percentage of Strains with National Center for Biotechnology Information
| Strain Code | 16s rRNA Sequence | Identification | Similarity, % |
|---|---|---|---|
| Strain aA | 1496 nucleotides | Pseudomonas aeruginosa JCM 5962(T) | 100 |
| Strain bB | 1516 nucleotides | Staphylococcus haemolyticus MTCC 3383(T) | 99.86 |
| Strain cC | 1484 nucleotides | Micrococcus yunnanensis YIM 65004(T) | 99.86 |
| Strain dD | 1492 nucleotides | Rothia amarae JCM 11375(T) | 99.79 |
| Strain eE | 1483 nucleotides | Gordonia terrae NBRC 100016(T) | 100 |
| Strain fF | 1495 nucleotides | Brevibacillus agri DSM 6348(T) | 99.93 |
5. Discussion
The present study showed that each salt medium provided growth conditions for some of the microorganisms found in paint sludge. Micrococcus yunnanensis was isolated from paint sludge in MSM, while in the same medium, Phulpoto et al. (27) isolated Brevibacillus parabrevis strains from oil-based paint sludge and Ashwini et al. (28) isolated Pseudomonas, Staphylococcus, and Lactobacillus stains. In the direct and serial culture of paint sludge on the NA medium, bacteria, including Rothia amarae, Gordonia terrae, and Brevibacillus agri were isolated. Obidi et al. (29) could successfully isolate Bacillus brevis, B. laterosporus, B. polymyxa, Escherichia coli, Lactobacillus gasser, Lactobacillus brevis, and Proteus mirabilis from fresh industrial paint on the NA medium and Aspergillus on the PDA medium. In the current study, no fungal colonies and Acidithiobacillus strains were isolated on PDA and 9k medium, respectively.
The Bushnell Haas medium that was applied to cultivate hydrocarbon-degrading microorganisms showed the most suitable growth of the species. Further, only Pseudomonas bacteria grew on the Bushnell Haas medium. The microbial population of paint sludge was counted as 1 × 104 CFU/mL on the PCA medium, whereas Maduka et al. (30) isolated microorganisms between 0 and 5.4 × 105 CFU/mL from emulsion paints and between 0 and 6.5 × 105 CFU/mL from clear paints. The isolated species were mostly pathogens that were isolated from clinical samples and soils, but the strains were different from previous studies. The bacteria Micrococcus yunnanensis, Staphylococcus haemolyticus, and Pseudomonas aeruginosa were isolated from the paint sludge. The sequences compared to the NCBI were 99.86% and 100% similar to YIM 65004, MTCC 3383, and JCM 5962 type strains, respectively. Opperman and Goll (31) isolated Bacillus, Pseudomonas, Proteus, Serratia, and Enterobacter as Gram-negative bacteria and Micrococcus and Clostridium as Gram-positive strains from water-based paints. Cappitelli et al. (32) isolated Staphylococcus from surfaces coated with epoxy resins, which was examined in terms of bioremediation. Brevibacillus agri isolated from the paint sludge had a 99.93% similarity to the DSM strain. Phulpoto et al. (27) in a study on paint bioremediation could isolate Brevibacillus parabrevis strain NAP3 from solvent-based paint. Rothia amarae strain was isolated from the paint sludge with 99.79% similarity to JCM 11375 strain but this bacterium was isolated from wastewater sludge by Fan et al. (33). In this study, Gordonia terrae strain with 100% similar to NBRC 100016 strain isolated from paint sludge. Recently, many Gordonia strains have isolated from various medium sources, such as soil, some of which were involved in the degradation of xenobiotic compounds, such as benzopyridine (34).
5.1. Conclusions
According to strains obtained in this study and the presence of resistant microorganisms in the automotive paint sludge, one can investigate the idea of the bioremediation using microorganisms existing in this type of sludge to reduce the pollution load and make the compliance with landfill standards.
Acknowledgements
References
-
1.
Bentley J, Turner GPA. Introduction to paint chemistry and principles of paint technology. USA: CRC Press; 1998. https://doi.org/10.1007/978-1-4899-3180-1.
-
2.
Talbert R. Paint technology handbook. USA: CRC Press; 2007. https://doi.org/10.1201/9781420017786.
-
3.
Ulbrich M, Kalendova A. Properties of organic coatings with nonisometric ferrite particles. Phys Proced. 2013;44:247-55. https://doi.org/10.1016/j.phpro.2013.04.030.
-
4.
Ravikumar HR. Biodegradation of paints: A current status. Indian J Sci Technol. 2012;5(1):1-11. https://doi.org/10.17485/ijst/2012/v5i1.33.
-
5.
Dalmazzo D, Vercelli A, Santagata E, Ruffino B, Zanetti MC. Rheological characterization and performance-related evaluation of paint sludge modified binders. Mater Struct. 2016;50(1). https://doi.org/10.1617/s11527-016-0945-y.
-
6.
Salihoglu G, Salihoglu NK. A review on paint sludge from automotive industries: Generation, characteristics and management. J Environ Manage. 2016;169:223-35. [PubMed ID: 26773426]. https://doi.org/10.1016/j.jenvman.2015.12.039.
-
7.
Zakaie F, Jafari A, Harirchian S. Feasibility of using the products recycled from the sludge in painting salons as a concrete and asphalt supplement. The First Specialized Conference on Environmental Engineering. Tehran. Department of Environment, University of Tehran; 2006.
-
8.
Burande BC. Utilisation of paint sludge from automotive industries into valueable products. Int J Recent Trend Engin Res. 2017;3(5):513-9.
-
9.
Khezri SM, Abdollahi F. Recycling of materials from the automotive paint sludge industry. The 4th Specialized Conference on Environmental Engineering. Tehran. University of Tehran. Department of Environment; 2010.
-
10.
Goldstein AH, Galbally IE. Known and unknown organic constituents in the Earth' s atmosphere. Environ Sci Technol. 2007;41(5):1514-21. [PubMed ID: 17396635]. https://doi.org/10.1021/es072476p.
-
11.
Salihoglu NK, Ucaroglu S, Salihoglu G. Bioconversion of industrial wastes: Paint sludge from automotive manufacturing. J Mater Cycles Waste Manage. 2018;20(4):2100-9. https://doi.org/10.1007/s10163-018-0764-z.
-
12.
Ghomi Avili R, Takdastan A, Atabi F, Omrani GA. Feasibility study of chromium removal from paint sludge with biological sludge, using vermicompost by eisenia fetida (case study: Saipa automotive industry). Jundishapur Health Sci. 2018;10(3). https://doi.org/10.5812/jjhs.78891.
-
13.
Gillatt J. Bacterial and fungal spoilage of waterborne formulations during production and on storage: remedial and preventative measures. Surf Coat Int. 1992;75:387.
-
14.
Abatenh E, Gizaw B, Tsegaye Z, Wassie M. The role of microorganisms in bioremediation- a review. Open J Environ Biol. 2017;2(1):38-46. https://doi.org/10.17352/ojeb.000007.
-
15.
Piemonte V, Francioni F, Capocelli M, Prisciandaro M. Biodegradation of acrylic paints: Process modelling of biocide effect on biomass growth at different temperatures. Braz J Chem Engin. 2017;34(2):557-66. https://doi.org/10.1590/0104-6632.20170342s20150766.
-
16.
Borah D, Yadav RNS. Optimization of BH medium for efficient biodegradation of diesel, crude oil and used engine oil by a newly isolated bacillus cereus strain DRDU1 from an automobile engine. Biotechnology(Faisalabad). 2014;13(4):181-5. https://doi.org/10.3923/biotech.2014.181.185.
-
17.
HiMedia Laboratories. 2020. Available from: Himedialabs.com.
-
18.
Silverman MP, Lundgren DG. Studies on the chemoautotrophic iron bacterium Ferrobacillus ferrooxidans. I. An improved medium and a harvesting procedure for securing high cell yields. J Bacteriol. 1959;77(5):642-7. [PubMed ID: 13654231]. [PubMed Central ID: PMC290434]. https://doi.org/10.1128/JB.77.5.642-647.1959.
-
19.
Nakamura K, Miki H, Amano Y. Cell growth and accumulation of Thiobacillus thiooxidans S3 in a pH-controlled thiosulfate medium. J General Appl Microbiol. 1990;36(6):369-76. https://doi.org/10.2323/jgam.36.369.
-
20.
Afifi A, Motamedi H, Alizadeh B, Leilavi H. Isolation and identification of oil degrading bacteria from oil sludge in Abadan oil refinery. Environ Exp Biol. 2015;13:13-8.
-
21.
Joseph PJ, Joseph A. Microbial enhanced separation of oil from a petroleum refinery sludge. J Hazard Mater. 2009;161(1):522-5. [PubMed ID: 18468790]. https://doi.org/10.1016/j.jhazmat.2008.03.131.
-
22.
Nwachukwu SCU, Akpata TVI. Isolation of microorganisms by spread plate technique. Lagos: Principles of quantitative microbiology. University of Lagos Press; 2003.
-
23.
Bergey DH, Breed RS, Murray EGD, Hitchens AP. Manual of determinative bacteriology. 5th ed. CABI Direct; 1939.
-
24.
Adli M, Hassanshahian M, Krimi Niki A. [Isolation, identification and characterization of two species of Shyvanlay biosurfactant producing Persian Gulf]. J Microbe Word. 2013;6:53-61. Persian.
-
25.
Kim OS, Cho YJ, Lee K, Yoon SH, Kim M, Na H, et al. Introducing EzTaxon-e: A prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int J Syst Evol Microbiol. 2012;62(Pt 3):716-21. [PubMed ID: 22140171]. https://doi.org/10.1099/ijs.0.038075-0.
-
26.
EzTaxon server. Taxonomy. 2019. Available from: http://www.ezbiocloud.net/eztaxon.
-
27.
Phulpoto A, Qazi M, Mangi S, Ahmed S, Haq I, Phul A, et al. Bioremediation of oil-based paint from aqueous media by novel indigenous Brevibacillus parabrevis strain NAP3 and its toxicity assessment. Polish J Environ Stud. 2017;26(1):229-37. https://doi.org/10.15244/pjoes/62905.
-
28.
Ashwini R, Anchana D. Bacterial degradation of paint. Int J Biol Res. 2018;3(1):180-4.
-
29.
Obidi OF, Aboaba OO, Makanjuola MS, Nwachukwu SCU. Microbial evaluation and deterioration of paints and paint-products. J Environ Biol. 2009;30(5):835.
-
30.
Maduka CM, Igwilo NC. Microorganisms survive in paints. Cur Anal Biotechnol. 2019;2:1-5.
-
31.
Opperman RA, Goll M. Presence and effects of anaerobic bacteria in water-based paints. J Coatings Technol. 1984;56:51-7.
-
32.
Cappitelli F, Principi P, Pedrazzani R, Toniolo L, Sorlini C. Bacterial and fungal deterioration of the Milan Cathedral marble treated with protective synthetic resins. Sci Total Environ. 2007;385(1-3):172-81. [PubMed ID: 17658586]. https://doi.org/10.1016/j.scitotenv.2007.06.022.
-
33.
Fan Y, Jin Z, Tong J, Li W, Pasciak M, Gamian A, et al. Rothia amarae sp. nov., from sludge of a foul water sewer. Int J Syst Evol Microbiol. 2002;52(Pt 6):2257-60. [PubMed ID: 12508895]. https://doi.org/10.1099/00207713-52-6-2257.
-
34.
Kampfer P, Martin K, Dott W. Gordonia phosphorivorans sp. nov., isolated from a wastewater bioreactor with phosphorus removal. Int J Syst Evol Microbiol. 2013;63(Pt 1):230-5. [PubMed ID: 22389287]. https://doi.org/10.1099/ijs.0.037093-0.