Abstract
Context:
Numerous studies have shown the high frequency and antibiotic-resistant patterns of Shigella species in different provinces of Iran. In this study, we performed a comprehensive review from 2000 to 2020 in Iran to describe the prevalence rate and antibiotic-resistant patterns of S. sonnei, S. dysenteriae, S. flexneri, and S. boydii.Evidence Acquisition:
We systematically searched the biomedical databases including Scopus, Google Scholar, PubMed, SID, and Web of Science for related articles published in English or Persian. Finally, out of 70 articles, 34 studies were included in the study.Results:
From 44,292 clinical specimens, 2,742 cases were introduced as positive samples for Shigella species in Iran during 2000-2020. Also, S. sonnei (n = 1484, 54.1%) was the predominant species in Iran, followed by S. flexneri (n = 1100, 40.1%), S. dysenteriae (n = 80,3%), and S. boydii (n = 78, 2.8%). These Shigella species showed maximum resistance to ampicillin (n = 1759, 64%-96%), cotrimoxazole (n = 1220, 87%-100%), nalidixic acid (n = 649, 10%-82%), trimethoprim-sulfamethoxazole (n = 459, 80%-98.5%), cefotaxime (n = 410, 53%-63%), and tetracycline (n = 386, 36%-94%). No resistances were found against imipenem, meropenem, cefoxitin, norfloxacin, levofloxacin, azithromycin, and amoxicillin. Also, 308 and 359 cases were introduced as multidrug resistance (MDR) and extended spectrum beta lactamase (ESBL) producing species, respectively.Conclusions:
Evaluation of endemic shigellosis and antibiotic-resistant patterns through epidemiological studies are necessary to promote infection control strategies. These data may be useful to avoid empirical treatment, revise treatment guidelines, and decrease antimicrobial resistance of Shigella spp. in human societies.Keywords
1. Context
Gastrointestinal infections caused by different enteric bacterial pathogens such as Escherichia and Shigella are the main public health threat worldwide. Shigella is a gram-negative and intracellular bacterium with the pathogenic subgroup (A-D) with different distribution in developing countries. Shigella species belong to the Enterobacteriaceae family, which includes S. dysenteriae, S. flexneri, S. boydii, and S. sonnei, with exclusive epidemiological features. They are spread through the fecal-oral route and produce acute infection in the intestine called shigellosis. However, this infection is mostly caused by S. sonnei in industrialized countries and S. flexneri in developing countries (1-3). Moreover, S. dysenteriae is known as an epidemic form of shigellosis in different countries. According to the estimations, 160 million disease cases and 600,000 deaths are due to Shigella infections annually worldwide. In Iran, the prevalence of Shigella infections is so variable geographically. For example, different studies have shown that the prevalence of diarrhea caused by Shigella is almost high in different Iranian cities such as Kashan, Tehran, Kerman, Zanjan, and Ahvaz. In late summer 2006, during the final stage of an outbreak of shigellosis in the Isfahan Province, a diarrheal outbreak appeared to be the result of shigellosis (1-5). Generally, shigellosis is related to insufficient sanitation, some environmental factors, person-to-person contact, especially in young children, contaminated water and foods, sexual activity, and traveling. Also, Shigella pathogenesis is related to various virulence factors located in the chromosome (such as invasion-associated locus (ial) and Shigella enterotoxin 1 genes (set1A and set1B)) and large virulent inv plasmids (for example; invasion plasmid antigen H (ipaH) and Shigella enterotoxin 2 gene). These factors are associated with dissemination from cell to cell and the watery phase of diarrhea in the epithelial cells (1-6). The frequency and antibiotic-resistant patterns of Shigella species are changing rapidly over time. So, antibiotic treatments are typically suggested to decrease the symptoms of Shigella infections (6-9). Selecting an appropriate antibiotic for the treatment of Shigella infections is necessary because multidrug resistance (MDR) species can appear from many mechanisms, such as a decrease in cellular permeability, extrusion of drugs by active efflux pumps, and overexpression of drug-modifying in Shigella genomes (10-13). For example, the appearance of extended-spectrum-β-lactamases (ESBLs) producing strains of Shigella spp. and developing resistance to different treatment recommendations such as sulfonamides, tetracycline, chloramphenicol, ampicillin, fluoroquinolones, and ceftriaxone or azithromycin, which are recommended by the World Health Organization (WHO) for fluoroquinolones resistant species, are reported in different research studies worldwide. According to the WHO, effective treatment for Shigella spp. must be selected by the prevalence and antimicrobial susceptibility patterns of the endemic strains. Furthermore, some novel therapeutic strategies for Shigella treatment were suggested. For example, nanoparticles (NPs) have shown high antibacterial activity during in-vitro and in-vivo experiments, phage therapy, biotherapeutic agents (preferably probiotics), and natural and organic products (6, 10, 14). Today, different phenotypic and molecular methods have been used to diagnose Shigella species in human clinical specimens (11, 12, 14-16). Also, for epidemiological investigations, amplifying and non-amplifying DNA fingerprinting methods have been used for pathogenic strains. In Iran, different studies showed the prevalence and antimicrobial-resistant patterns of Shigella species isolated from clinical specimens of the adult and pediatric patients. In this comprehensive review, we tried to characterize and summarize this information in Iran during 2000-2020.
2. Evidence Acquisition
2.1. Search Strategy
We systematically searched the biomedical databases including Scopus, Google Scholar, PubMed, SID, and Web of Sciences in English and Persian to find the related studies from 2000 to 2020 with different keywords including “Shigella spp. AND Iran”, “Shigella dysenteriae AND Iran”, “Shigella flexeneri AND Iran”, “Shigella boydii AND Iran”, “Shigella sonnei AND Iran”, “Shigella spp. AND human clinical specimens AND Iran”, “Shigella spp. AND antimicrobial resistance AND Iran”, “Shigella dysenteriae OR Shigella flexeneri AND antimicrobial resistance AND Iran”, “Shigella boydii OR Shigella sonnei AND Iran”, and “Shigella spp. AND multidrug resistance (MDR), and ESBLs AND Iran”. To clarify the prevalence and development of antibiotic-resistance, we reviewed the published literature and their references to provide and categorize information about the prevalence and antimicrobial resistance of Shigella species isolated from different pediatrics and adult patients. Finally, out of 70 articles, we selected 34 papers published from 2000 to 2020 (Figure 1).
Flowchart of search strategies
2.2. The Inclusion Criteria
There were four inclusion criteria:
1) Epidemiological and frequency studies were selected and categorized based on year, type of human clinical specimen, age groups, and the frequency of different Shigella species. The data was separated by the province and regions, sample collection, sample size, and number of positive specimens for different Shigella species.
2) Research studies with different phenotypic and molecular methods such as stool culture, biochemical detection, serotyping tests, Multiplex PCR, PCR-RFLP, and PFGE assay to identify Shigella spp.
3) Different clinical specimens such as watery and bloody diarrhea and rectal swabs were collected from hospitalized patients with different clinical signs and symptoms.
4) The studies that focused on antimicrobial susceptibility tests (AST) for Shigella spp. according to clinical laboratory standard institute (CLSI) and performed through disk diffusion (Kirby-Bauer) or molecular methods (Tables 1 and 2).
Prevalence Studies for Shigella spp. Based on Clinical Human Specimens (ND: Not Detected)
| Reference, Publication Year | Performed Year | Region | Human Sample | Age, y; Male & Female | Detection Methods | Sample Size | Positive for Shigella spp., No. (%) | Frequency of Shigella spp., No. (%) | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| S. flexneri | S. sonnei | S. boydii | S. dysenteriae | ||||||||
| Salimiyan Rizi et al., 2020 (17) | 2018-2019 | Mashhad | Stool samples | < 14 | Culture, biochemical, serotyping tests. | 233 | 89 (22.3) | 22 (23.4) | 66 (70.2) | ND | 1 (1) |
| Karimi-Yazdi et al., 2020 (8) | 2017-2018 | Tehran | Diarrheal stool | ≤ 14 | Culture, PCR, slide agglutination | 141 | 141 (100) | 28 (19.9) | 111 (78.7) | 2 (1.4) | ND |
| Sheikh et al., 2019 (18) | 2016-2017 | Ahvaz | Bloody, mucoid and watery diarrhea | 2-65 | Culture, PCR, Slide agglutination | 522 | 69 (13.2) | 34 (49.3) | 22 (31.9) | 9 (13) | 4 (5.8) |
| Avakh Majalan et al., 2018 (6) | 2016 | Tehran | diarrheal stool | Randomly | Culture, biochemical, serotyping tests. | 300 | 26 (8.7) | 11 (42.3) | 15 (57.7) | ND | ND |
| Teimourpour et al., 2019 (13) | 2015-2017 | Ardabil | Stool samples | < 10 | Culture, PCR | 1280 | 113 | 22 (19.4) | 79 (69.9) | 9 (9.9) | 3 (6.2) |
| Moghanloo et al., 2018 (19) | 2015-2017 | Kashan | Diarrheal stool | 1-69 | Culture, biochemical, serotyping tests. | 528 | 98 (18.5) | 31 (31.63) | 57 (58.1) | 2 (2) | 8 (8.1) |
| Yaghoubi et al., 2017 (7) | 2015-2016 | Tehran | Stool samples | 1-15 | Culture, serotyping tests. Multiplex PCR | 946 | 75 (7/7) | 33 (44);1a (11), 2a (22) | 40 (53.3) | 1 (1.3) | 1 (1.3) |
| Shahin et al., 2020 (20) | 2015-2016 | Isfahan, Fars, Hormozgan, Kohkiloyeh va Boyer Ahmad | Not define | Not define | Biochemical, PCR, serogroup test | 70 | 70 (100) | 26 (37.1) | 44 (62.8) | ND | ND |
| Beladi Ghannadi et al., 2019 (21) | 2015-2016 | Tehran | Stool samples | Randomly | Culture, serotyping tests. Multiplex PCR | 945 | 52 (5.5) | 12 (23) | 23 (44.2) | 5 (9.6) | 12 (23) |
| Pour et al., 2016 (22) | 2015 | Ahvaz | Diarrheal stool | ≤ 2-8 | Culture, biochemical, ERIC-PCR | 50 | 50 | 31 (62) | 16 (32) | 3 (6) | ND |
| Yousefi et al., 2018 (23) | 2015 | Kerman | Stool samples | 15-79 | Culture, biochemical | 241 | 5 (19) | ND | ND | ND | 5 (19) |
| Abbasi et al., 2019, (2) | 2015 | Arak | Mucoid and bloody diarrhea | 1-10 | Culture, PCR, slide agglutination | 230 | 19 (8.2) | 4 (21) | 15 (78.9) | ND | ND |
| Shokoohizadeh et al., 2017 (24) | 2015 | Ahvaz | Diarrheal stool samples | ≤ 2-8 | Microbiologic tests, ERIC-PCR, MLST | 80 | 50 (62.5) | 31 (62) | 16 (32) | 3 (6) | ND |
| Aminshahidi et al., 2017 (3) | 2014-2015 | Shiraz | Loose stools with WBC + | ≤ 18 | Culture, PCR | 269 | 41 (15.2) | 33 (80.5) | 8 (19.5) | ND | ND |
| Soltan Dallal et al., 2015 (25) | 2013-2014 | Tehran | Stool samples | ≤ 14 | Culture, biochemical test | 200 | 6 (3) | ND | 6 (3) | ND | ND |
| Hosseini Nave et al., 2016 (26) | 2013-2014 | Kerman | Stool samples | Randomly | Biochemical, serological test, Multiplex PCR | 624 | 56 (9) | 31 (55.4) | 18 (32.1) | 7 (12.5) | ND |
| Talebreza et al., 2015 (27) | 2013-2014 | Tehran | Stool samples | < 10 | Culture, PCR, slide agglutination | 938 | 36 | 10 (27.8) | 22 (61.1) | 3 (8.3) | 1 (2.8) |
| Nikfar et al., 2017 (4) | 2013-2014 | Ahvaz | Stool samples | ≤ 12 | Culture, slide agglutination | 193 | 193 (100) | 125 (64.8) | 63 (32.6) | 4 (2.1) | 1 (0.5) |
| Nodeh Farahani et al., 2018 (28) | 2012-2016 | Tehran | stool samples | 1-10 | Culture, slide agglutination | 5300 | 472 (8.9) | 185 (39.2) | 287 (60.8) | ND | ND |
| Zahedi Bialvaei et al., 2016 (16) | 2012-2013 | Tehran, Fars, Kurdistan, Mazandaran, Khuzestan, Sistan va Baluchestan | Urine, blood, sputum, wound, respiratory and vaginal secretions, biopsies, body fluids | Randomly | Culture, biochemical test, slide agglutination | 443 | 52 (11/7) | ND | ND | ND | ND |
| Alizadeh-Hesar et al., 2015 (11) | 2012-2013 | Tehran | Bloody and loose stools | < 5 | Culture, PCR, PFGE | 5291 | 70 (1.32) | 8 (11.43) | 61 (87.14) | 1 (1.43) | ND |
| Jomezadeh et al., 2014 (1) | 2011-2013 | Abadan | Stool samples | ≤ 1-15 | Culture, biochemical test, Slide agglutination | 705 | 36 (5.1) | 19 (52.7) | 11 (30.5) | 4 (11.1) | 2 (5.5) |
| Mostafavi et al., 2016 (29) | 2010-2015 | Isfahan | Stool samples | < 5-15 < | Culture, biochemical test, slide agglutination | 45 | 45 (100) | 15 (34.1) | 28 (63.6) | ND | 1 (2.3) |
| Zahedi Bialvaei et al., 2017 (30) | 2009-2013 | Tabriz | Stool samples | 3-70 | Culture, biochemical test, Slide agglutination, PCR | 58 | 58 (100) | 7 (12) | 45 (77.6) | ND | 6 (10.3) |
| Esmaeili Dooki et al., 2014 (31) | 2009 | Mazandaran | Fecal specimen and Rectal swab | ≤ 1-14 | Culture, biochemical test | 1072 | 7 (0.65) | ND | 6 (85) | ND | 1 (15) |
| Ranjbar et al., 2013 (5) | 2008-2010 | Tehran | Rectal swabs | ≤ 12 | Culture, biochemical test, Slide agglutination | 55 | 55 (100) | ND | ND | ND | ND |
| Ranjbar and Mirsaeed Ghazi, 2013 (32) | 2008-2010 | Tehran | Rectal swabs | ≤ 5-12 ≤ | Culture, biochemical test, ERIC-PCR | 950 | 89 (9.3) | 28 (31.5) | 54 (60.7) | 5 (5.6) | 2 (2.2) |
| Khaghani et al., 2014 (9) | 2008-2010 | Ahvaz | Stool samples | ≤ 1-12 | Culture, biochemical test, slide agglutination | 4380 | 175 (4) | 87(49.8); type 1=19(21.8), type 2=50 (57.5), type 3=1 (1), type 4=3(3.4), type 6=7(8.1) | ND | ND | ND |
| Ranjbar and Memariani, 2015 (33) | 2008-2010, 2002-2003 | Tehran | Watery, loose and bloody stools | ≤ 12 | Culture, slide agglutination, MLVA assay | 950 | 47 (4.9) | ND | 47 (100) 21 genotypes | ND | ND |
| Dibaj et al., 2013 (34) | 2006 | Isfahan | Rectal swabs | Randomly | Culture, biochemical test, slide agglutination, Ribotyping | 146 | 13 (8.9) | 2 (15.4) | 6 (46.1) | 1 (7.7) | 4 (30.8) |
| Soltan Dallal et al., 2019 (35) | 2005-2006 | Esfahan, Tehran, Kurdistan, Yazd Ghazvin, Zanjan, Semnan, Golestan | Fecal swab samples | Randomly | Culture, serogroup test, PCR | 1012 | 29 (2.86) | 13 (44.8) | 16 (55.2) | ND | ND |
| Farshad et al., 2015 (36) | 2003 | Shiraz | Stool samples | ≤ 1-14 | PCR-RFLP and PFGE | 719 | 82 (11.4) | 16 (19.51) | 61 (74.3) | 3 (3.6) | 2 (2.4) |
| Gharibi et al., 2012 (15) | 2002-2008 | Bushehr | Stool samples | Randomly | Culture, biochemical test, slide agglutination | 121 | 121 (100) | 46 (38.1) | 62 (51.2) | 8 (6.6) | 5 (4.1) |
| Pourakbari et al., 2010 (12) | 2001-2006 | Tehran | Stool samples | Randomly | Culture, biochemical test, slide agglutination | 15,255 | 397 (2.6) | 190 (47.9) | 179 (45.1) | 8 (2) | 20 (5) |
Characteristics of Antimicrobial Resistance Patterns of Shigella spp. in Studies Performed in Iran
| Antimicrobial Agents | Number of Studies | Resistant Strains Reported, Number (from 2742 Shigella spp.) | Rate of Resistance Reported, No. (%) | Reference |
|---|---|---|---|---|
| Ampicillin | 14 | 1759 | 64-96% | (1, 6, 8, 9, 12, 14, 17, 18, 20, 21, 25, 28, 30, 31) |
| Trimethoprim-sulfamethoxazole | 8 | 452 | 80-98.5% | (1, 3, 8, 9, 14, 18, 21, 30) |
| Chloramphenicol | 10 | 299 | 10-61% | (1, 2, 6, 9, 12, 14, 18, 20, 25, 30) |
| Nalidixic acid | 16 | 649 | 10-82% | (1-3, 6, 8, 9, 11-15, 17, 18, 20, 28, 31) |
| Gentamicin | 15 | 260 | 1.5-36% | (1-4, 6, 9, 11-14, 17, 18, 25, 30, 31) |
| Erythromycin | 2 | 51 | 57-68% | (18, 31) |
| Cefixime | 6 | 169 | 22-68% | (1, 2, 8, 17, 18, 31) |
| Ceftazidime | 6 | 217 | 27-39% | (2, 3, 12, 18, 21, 27) |
| Ceftriaxone | 10 | 311 | 0-63% | (1-4, 6, 13, 14, 17, 18, 31) |
| Ciprofloxacin | 14 | 105 | 0-40% | (1-3, 6, 8, 13-15, 18, 20, 21, 25, 31, 35) |
| Tetracycline | 11 | 386 | 36-94% | (1, 2, 6, 11, 14, 15, 20, 21, 25, 27, 30) |
| Cotrimoxazole | 7 | 1220 | 87-100% | (2, 11, 12, 15, 17, 25, 28) |
| Cefotaxime | 10 | 410 | 53-63% | (2, 3, 8, 14, 20, 25, 27, 28, 30, 31) |
| Ceftizoxime | 5 | 74 | 6-41% | (2, 12, 15, 30, 31) |
| Cefoxitin | 1 | 3 | 15% | (2) |
| Norfloxacin | 2 | 6 | 4-5% | (2, 14) |
| Azithromycin | 4 | 131 | 0-47% | (2, 3, 13, 17) |
| Imipenem | 2 | 0 | 0 | (2, 13) |
| Meropenem | 1 | 0 | 0 | (3) |
| Amikacin | 6 | 176 | 0-63% | (3, 12, 13, 21, 30, 31) |
| Levofloxacin | 1 | 16 | 11% | (8) |
| Minocycline | 1 | 158 | 66-93% | (8) |
| Streptomycin | 3 | 144 | 98-100% | (11,21,36) |
| Tobramycin | 1 | 112 | 20% | (12) |
| Kanamycin | 1 | 358 | 60% | (12) |
| Cephalothin | 1 | 311 | 42-67% | (12) |
| Aztreonam | 1 | 19 | 34% | (14) |
| Ofloxacin | 2 | 8 | 4-5% | (13, 14) |
| Amoxicillin | 2 | 30 | 40-83% | (17, 27) |
| ESBL producing | 5 | 359 | 5.7-56% | (2, 3, 5, 16, 28) |
| MDR | 6 | 308 | 76-98% | (2, 5, 6, 8, 9, 21) |
2.3. Exclusion Criteria
Case reports, duplicate documents, and the studies not exploring the prevalence and antimicrobial resistance for S. sonnei, S. flexneri, S. dysenteriae, and S. boydii were excluded from the study.
2.4. Data Analysis
In this review, data extraction was performed independently by three researchers to contradict any possibility of error. We used Microsoft Excel 2019 for storage, statistical analyses, numerical calculations, and chart designing. Also, for continuous variables, Wilcoxon rank-sum test (P-value ≤ 0.05) was used.
3. Results
3.1. Prevalence Rates for Pathogenic Shigella Species
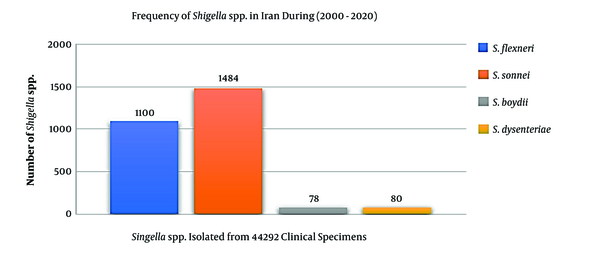
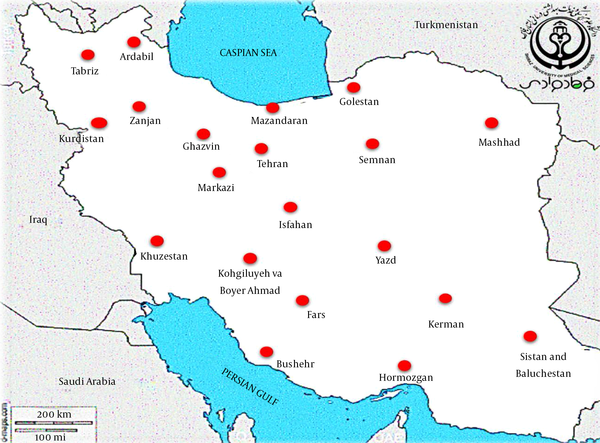
The epidemiological information and frequency of different Shigella spp. were categorized and presented in Table 1. In total, 44,292 human clinical specimens to find Shigella species in different provinces of Iran during 2000-2020 were investigated. Most studies (n = 12) had been performed during 2013-2016, and nine studies had been performed in 2015. We selected six observational cohort studies that were performed during five years. According to the collected data, 2,742 clinical sample cases were introduced as a positive sample for the Shigella species (Table 1). The frequencies of the Shigella dysenteriae (subgroup A), Shigella flexeneri (subgroup B), Shigella boydii (subgroup C), and Shigella sonnei (subgroup D) in Iran were compared (Figure 2). Furthermore, these studies were done in different provinces such as Tehran, Isfahan, Fars, and Mazandaran in Iran. According to different reports, most of the Shigella infections were discovered in Tehran (capital of Iran), Abadan, Isfahan, and Fars provinces. The geographic distribution of the prevalence studies in different provinces is shown in Figure 3. In these studies, different clinical stool specimens such as watery, bloody, and mucoid diarrhea and rectal swabs were collected from adult and pediatric age groups. Also, many of these specimens were isolated from hospitalized patients with different signs and symptoms. Some studies were defined in primary clinical outcome (i.e. diarrhea, fever, and abdominal cramps) in children. Also, the major manifestations of the disease for each species and strain were bloody, mucoid, and watery diarrhea. One study had been performed in six provinces of Iran and used other different samples such as urine, sputum, wound, respiratory fluids, vaginal secretions, biopsies, and blood culture. All isolates were confirmed as Shigella species by microbiological methods (e.g. culture, biochemical and serological tests by slide agglutination and a group-specific polyvalent antiserum) and molecular methods (e.g. Multiplex PCR, ERIC-PCR, and PCR-RFLP). Some studies used multilocus variable-number tandem-repeat (VNTR) analysis (MLVA) for genotyping of local Shigella strains. Also, the major phenotypic and genotypic methods to identify Shigella species were culture, biochemical test, slide agglutination, and Multiplex PCR. Out of 2,742 positive samples for Shigella species, S. sonnei (n = 1,484, 54.1%) was a predominant species in Iran, followed by S. flexneri (n = 1,100, 40.1%), S. dysenteriae (n = 80, 3%), and S. boydii (n = 78, 2.8%). Some of the studies reported the prevalence and relations between Shigella virulence genes and shigellosis in Iran. They reported some virulence genes such as ipaH, ipaBCD (necessary for invasion and intracellular survival), VirA (intracellular spreading factor), stx, set1A, set1B, and sat among Shigella species in pediatric or hospitalization diarrhea. For example, one of the studies showed the prevalence of enterotoxins ShET-2 (sen), ipaH, ipaBCD, sat, virA, ial, set1A, and set1B genes in Shigella species isolated from hospitalizing bloody diarrhea or other study revealed the high prevalence of ipaH, ipaC, sen, ipaD, virA, ipaB, and ipgD genes in Shigella isolates (7, 8).
The frequency of the Shigella dysenteriae (subgroup A), Shigella flexeneri (subgroup B), Shigella boydii (subgroup C), and Shigella sonnei (subgroup D) in Iran during 2000-2020
The geographic distribution of the prevalence studies for Shigella spp. in different provinces of Iran during 2000 -2020
3.2. Antibiotic-Resistance Patterns of Shigella Species
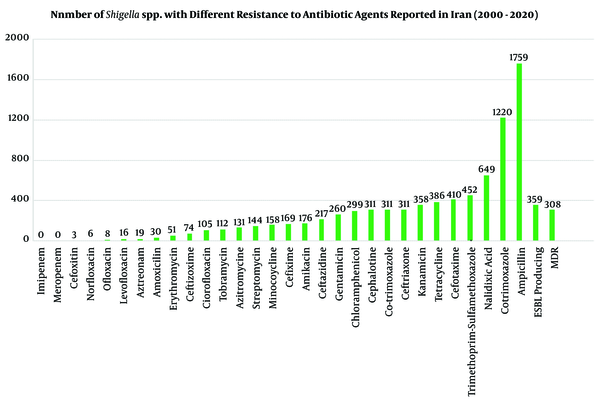
The antibiotic-resistance information for positive samples (n = 2,742 Shigella species) against 30 most common antibiotic agents was investigated in Iran during 2000-2020. The AST for Shigella isolates was completed by phenotypic according to CLSI guidelines and genotypic methods. In different studies, Shigella isolates (n = 2,742) showed maximum resistance to Ampicillin (n = 1,759, 64%-96%), Cotrimoxazole (n = 1,220, 87%-100%), Nalidixic acid (n = 649, 10%-82%), Trimethoprim-sulfamethoxazole (n = 459, 80%-98.5%), Cefotaxime (n = 410, 53%-63%), and Tetracycline (n = 386, 36%-94%). No resistances were found against imipenem, meropenem, cefoxitin, norfloxacin, levofloxacin, azithromycin, and amoxicillin. Furthermore, MDR phenotypes were seen in six studies for 308 Shigella species with different resistance patterns to antibiotic agents. Also, 359 Shigella isolates in five studies could produce ESBLs, and some of them were positive for blaTEM, blaCTX-M-1, blaCMY 2, blaCIT, and bla CTX-M-15 (Table 2 and Figure 4) (2, 3, 5, 16, 28).
Antibacterial resistant patterns of Shigella spp. in Iran during 2000-2020
4. Discussion
Several studies reported that the prevalence and antibiotic-resistant patterns of Shigella spp. are on the rise in Iran. According to the WHO, exploring the epidemiology of the infectious disease in developing countries is necessary because annual reports have shown that about 200,000 infectious diseases are related to this Shigella spp. Hence, to explain the prevalence and antibiotic-resistant patterns of Shigella spp., we reviewed the related studies published from 2000 to 2020 in Iran. Overall, Shigella virulence or pathogenicity is related to the immunity of patients, and severe clinical findings with low infectious dose (10-100 organisms) can be seen in children, elderly adults, and immunodeficient patients. This bacterial species is transmitted by the oral-fecal pathway and causes self-limiting diarrhea or invasive bacillary dysentery with bleeding or inflammatory diarrhea, fever, and abdominal cramps. Unfortunately, antibiotic therapy is a main strategy to combat and control the Shigella spread, and excessive use of antibiotics agents can develop MDR strains in different countries. According to the Centers for Disease Control and Prevention (CDC), Shigella infections are treated with ciprofloxacin and Ceftriaxone, especially for children with shigellosis. Hence, understanding the prevalence and antibiotic-resistance patterns of Shigella species is necessary for efficient treatment and increasing the public hygiene (17, 22, 34, 36-39). Different countries such as Bangladesh, Maldives, Tanzania, Nepal, Myanmar, and Sri Lanka reported the prevalence of Shigella spp. in symptomatic and asymptomatic children (40). In Iran, different studies were performed to evaluate the prevalence and antibacterial resistance patterns for Shigella subgroup (A-D) in pediatric and adult patients. For example, in Thailand and United States, S. sonnei had the highest prevalence, followed by S. flexneri during 1997-2006. Similarly, our comprehensive review showed the S. sonnei (54.1%) was a predominant species in Iran during a 20 year period. But other studies in China, Bangladesh, Pakistan, Indonesia, Nepal, and Vietnam documented S. flexneri as the foremost species in these regions (33, 41-44). Furthermore, various studies documented the S. sonnei and S. flexneri outbreaks in Maharashtra, West Bengal, and Kerala (26). In addition, some of the studies showed different antibiotic-resistance patterns for Shigella species in Iran and other countries. For instance, according to different reports in Iran, Shigella species have a maximum resistance to ampicillin, cotrimoxazole, nalidixic acid, trimethoprim-sulfamethoxazole, cefotaxime, and tetracycline. compared to other countries, resistance to ciprofloxacin, amoxicillin, and cotrimoxazole was detected in Shigella species in Pakistan, Bangladesh, Vietnam, and China (33, 42, 43). Some of these antibiotics agents were described in the National Antimicrobial Resistance Monitoring system (NARMS 2015) Report for Shigella species. The NARMS introduces Shigella as an important MDR phenotype, including resistance to at least ampicillin, trimethoprim-sulfamethoxazole, tetracycline, and sulfisoxazole. These antibiotics and the results from antibiotic resistance are similar to AST platforms performed according to the CLSI in Iranian clinical research. Also, for Shigella, fluoroquinolones and macrolides are important agents in the treatment of severe infections (33, 44). Moreover, different studies (for example, in Chandigarh from 2001 to 2009 and Korea from 1991 to 2002) reported a high level of ESBL positive in Shigella strains (26, 33, 42-44). Also, in Iran, 11 studies introduced 667 clinical isolates as MDR and ESBLs producing species.
4.1. Some Information About
Shigella infections in Iran is still unknown as follows: prevalence rate, predominant species, and antibacterial resistant studies from all provinces, role of Shigella virulence factors in various infections, and type of experimental treatment of the shigellosis in different provinces. Also, understanding the resistance and susceptibility to different antibiotics for Shigella species may assist in revising treatment procedures and develop an active treatment for shigellosis in pediatric and adult patients. In this study, we determined the prevalence of different Shigella species, predominant species, the provinces that have an important role in Shigella detections, and the main laboratory diagnostic methods in Iran. We also introduced the maximum and minimum resistance to different types of antibiotics in Iran for 20 years. According to different studies in Iran, it is not possible to conclude the main sources of infection in Iran, the relations between Shigella virulence factors in various infections, and the main transmission mechanisms responsible for antibiotic resistance.
5. Conclusions
Evaluation of endemic shigellosis through epidemiological studies is necessary to define the source of infection and promote infection control policies. Future studies in Iran should determine the prevalence, antibiotics resistance rates, sources of infection, virulence factors in various infections, and transmission of the antibiotic resistance mechanisms of Shigella spp. in all provinces. This data can be useful to avoid empirical therapy, choose the best antibiotics for effective treatment, and improv public health in human society.
References
-
1.
Jomezadeh N, Babamoradi S, Kalantar E, Javaherizadeh H. Isolation and antibiotic susceptibility of Shigella species from stool samples among hospitalized children in Abadan, Iran. Gastroenterol Hepatol Bed Bench. 2014;7(4):218-23. [PubMed ID: 25289136]. [PubMed Central ID: PMC4185876].
-
2.
Abbasi E, Abtahi H, van Belkum A, Ghaznavi-Rad E. Multidrug-resistant Shigella infection in pediatric patients with diarrhea from central Iran. Infect Drug Resist. 2019;12:1535-44. [PubMed ID: 31239729]. [PubMed Central ID: PMC6559769]. https://doi.org/10.2147/IDR.S203654.
-
3.
Aminshahidi M, Arastehfar A, Pouladfar G, Arman E, Fani F. Diarrheagenic Escherichia coli and Shigella with High Rate of Extended-Spectrum Beta-Lactamase Production: Two Predominant Etiological Agents of Acute Diarrhea in Shiraz, Iran. Microb Drug Resist. 2017;23(8):1037-44. [PubMed ID: 29106325]. [PubMed Central ID: PMC5709696]. https://doi.org/10.1089/mdr.2017.0204.
-
4.
Nikfar R, Shamsizadeh A, Darbor M, Khaghani S, Moghaddam M. A Study of prevalence of Shigella species and antimicrobial resistance patterns in paediatric medical center, Ahvaz, Iran. Iran J Microbiol. 2017;9(5):277-83. [PubMed ID: 29296272]. [PubMed Central ID: PMC5748446].
-
5.
Ranjbar R, Mirsaeed Ghazi F, Farshad S, Giammanco GM, Aleo A, Owlia P, et al. The occurrence of extended-spectrum beta-lactamase producing Shigella spp. in Tehran, Iran. Iran J Microbiol. 2013;5(2):108-12. [PubMed ID: 23825726]. [PubMed Central ID: PMC3696844].
-
6.
Avakh Majalan P, Hajizade A, Nazarian S, Pourmand MR, Siyavoshani KA. Investigating the Prevalence of Shigella Species and Their Antibiotic Resistance Pattern in Children With Acute Diarrhea Referred to Selected Hospitals in Tehran, Iran. J Appl Biotechnol Rep. 2018;5(1):70-4. https://doi.org/10.29252/jabr.05.02.06.
-
7.
Yaghoubi S, Ranjbar R, Dallal MMS, Fard SY, Shirazi MH, Mahmoudi M. Profiling of Virulence-associated Factors in Shigella Species Isolated from Acute Pediatric Diarrheal Samples in Tehran, Iran. Osong Public Health Res Perspect. 2017;8(3):220-6. [PubMed ID: 28781945]. [PubMed Central ID: PMC5525559]. https://doi.org/10.24171/j.phrp.2017.8.3.09.
-
8.
Karimi-Yazdi M, Ghalavand Z, Shabani M, Houri H, Sadredinamin M, Taheri M, et al. High Rates of Antimicrobial Resistance and Virulence Gene Distribution Among Shigella spp. Isolated from Pediatric Patients in Tehran, Iran. Infect Drug Resist. 2020;13:485-92. [PubMed ID: 32104018]. [PubMed Central ID: PMC7025676]. https://doi.org/10.2147/IDR.S238559.
-
9.
Khaghani S, Shamsizadeh A, Nikfar R, Hesami A. Shigella flexneri: a three-year antimicrobial resistance monitoring of isolates in a Children Hospital, Ahvaz, Iran. Iran J Microbiol. 2014;6(4):225-9. [PubMed ID: 25802704]. [PubMed Central ID: PMC4367937].
-
10.
Rahman M, Shoma S, Rashid H, El Arifeen S, Baqui AH, Siddique AK, et al. Increasing spectrum in antimicrobial resistance of Shigella isolates in Bangladesh: resistance to azithromycin and ceftriaxone and decreased susceptibility to ciprofloxacin. J Health Popul Nutr. 2007;25(2):158-67. [PubMed ID: 17985817]. [PubMed Central ID: PMC2753991].
-
11.
Alizadeh-Hesar M, Bakhshi B, Najar-Peerayeh S. Clonal dissemination of a single Shigella sonnei strain among Iranian children during Fall 2012 in Tehran, I.R. Iran. Infect Genet Evol. 2015;34:260-6. [PubMed ID: 26117443]. https://doi.org/10.1016/j.meegid.2015.06.024.
-
12.
Pourakbari B, Mamishi S, Mashoori N, Mahboobi N, Ashtiani MH, Afsharpaiman S, et al. Frequency and antimicrobial susceptibility of Shigella species isolated in Children Medical Center Hospital, Tehran, Iran, 2001-2006. Braz J Infect Dis. 2010;14(2):153-7. [PubMed ID: 20563441]. https://doi.org/10.1590/s1413-86702010000200007.
-
13.
Teimourpour R, Babapour B, Esmaelizad M, Arzanlou M, Peeri-Doghaheh H. Molecular characterization of quinolone resistant Shigella spp. isolates from patients in Ardabil, Iran. Iran J Microbiol. 2019;11(6):496-501. https://doi.org/10.18502/ijm.v11i6.2221.
-
14.
Ranjbar R, Farahani A. Shigella: Antibiotic-Resistance Mechanisms And New Horizons For Treatment. Infect Drug Resist. 2019;12:3137-67. [PubMed ID: 31632102]. [PubMed Central ID: PMC6789722]. https://doi.org/10.2147/IDR.S219755.
-
15.
Gharibi O, Zangene S, Mohammadi N, Mirzaei K, Karimi A, Gharibi A, et al. Increasing antimicrobial resistance among Shigella isolates in the Bushehr, Iran. Pak J Biol Sci. 2012;15(3):156-9. [PubMed ID: 22866547]. https://doi.org/10.3923/pjbs.2012.156.159.
-
16.
Zahedi Bialvaei A, Kafil HS, Asgharzadeh M, Aghazadeh M, Yousefi M. CTX-M extended-spectrum beta-lactamase-producing Klebsiella spp, Salmonella spp, Shigella spp and Escherichia coli isolates in Iranian hospitals. Braz J Microbiol. 2016;47(3):706-11. [PubMed ID: 27268117]. [PubMed Central ID: PMC4927670]. https://doi.org/10.1016/j.bjm.2016.04.020.
-
17.
Salimiyan Rizi K, Farsiani H, Sasan MS. High rate of resistance to ceftriaxone and azithromycin among Shigella spp. isolates at three children's referral hospitals in Northeast Iran. J Infect Chemother. 2020;26(9):955-8. [PubMed ID: 32446727]. https://doi.org/10.1016/j.jiac.2020.04.022.
-
18.
Sheikh AF, Moosavian M, Abdi M, Heidary M, Shahi F, Jomehzadeh N, et al. Prevalence and antimicrobial resistance of Shigella species isolated from diarrheal patients in Ahvaz, southwest Iran. Infect Drug Resist. 2019;12:249-53. [PubMed ID: 30774392]. [PubMed Central ID: PMC6348961]. https://doi.org/10.2147/IDR.S187861.
-
19.
Moghanloo E, Khorshidi A, Badameh P, Ghadirian Abarghuei A, Valipour M, Akbari H. Prevalence of Shigella and Other Pathogenic Gram-negative Bacteria in the Patients with Diarrhea in Kashan City, Iran During 2015-2017. J Hum Environ Health Prom. 2018;4(3):106-10. https://doi.org/10.29252/jhehp.4.3.2.
-
20.
Shahin K, Bouzari M, Komijani M, Wang R. A New Phage Cocktail Against Multidrug, ESBL-Producer Isolates of Shigella sonnei and Shigella flexneri with Highly Efficient Bacteriolytic Activity. Microb Drug Resist. 2020;26(7):831-41. [PubMed ID: 32031907]. https://doi.org/10.1089/mdr.2019.0235.
-
21.
Beladi Ghannadi S, Ghane M, Babaeekhou L. Determination of Antibiotic Resistance Pattern and frequency of CTX-M, TEM, and SHV Β-Lactamase Encoding Genes among Shigella Isolates from Inpatients in Tehran, Iran. Med Lab J. 2019;13(2):8-15. https://doi.org/10.29252/mlj.13.2.8.
-
22.
Pour MBMG, Shokoohizadeh L, Navab-Akbar FT. Analysis of clonal relationships among Shigella spp. isolated from children with shigellosis in Ahvaz, Iran. Arch Adv Biosci. 2016;7(2):45-51.
-
23.
Yousefi M, Dehesh MM, Askarpour S, Saeidi R. A Food Poisoning Outbreak by Shigella boydii in Kerman-Iran. Arch Clin Infect Dis. 2018;13(6). 82350. https://doi.org/10.5812/archcid.82350.
-
24.
Shokoohizadeh L, Kaydani GA, Ekrami A. Molecular characterization of Shigella spp. isolates from a pediatric hospital in Southwestern Iran. Gastroenterol Hepatol Bed Bench. 2017;10(4):319-22. [PubMed ID: 29379598]. [PubMed Central ID: PMC5758741].
-
25.
Soltan Dallal MM, Eghbal M, Sharafianpour A, Zolfaghari MR, Yazdi MKS. Prevalence and multiple drug resistance of Shigella sonnei isolated from diarrheal stool of children. J Med Bacteriol. 2015;4(3-4):24-9.
-
26.
Hosseini Nave H, Mansouri S, Sadeghi A, Moradi M. Molecular diagnosis and anti-microbial resistance patterns among Shigella spp. isolated from patients with diarrhea. Gastroenterol Hepatol Bed Bench. 2016;9(3):205-10. [PubMed ID: 27458513]. [PubMed Central ID: PMC4947135].
-
27.
Talebreza A, Memariani M, Memariani H, Shirazi MH, Eghbali Shamsabad P, Bakhtiari M. Prevalence and Antibiotic Susceptibility of Shigella Species Isolated From Pediatric Patients in Tehran. Arch Pediatr Infect Dis. 2015;4(1). https://doi.org/10.5812/pedinfect.32395.
-
28.
Nodeh Farahani N, Masjedian Jazi F, Nikmanesh B, Asadolahi P, Sadeghi Kalani B, Amirmozafari N. Prevalence and antibiotic susceptibility patterns of salmonella and shigella species isolated from pediatric diarrhea in Tehran. Arch Pediatr Infect Dis. 2018;6(4). 57328. https://doi.org/10.5812/pedinfect.57328.
-
29.
Mostafavi N, Bighamian M, Mobasherizade S, Kelishadi R. Resistance of Shigella strains to extended-spectrum cephalosporins in Isfahan province. Med J Islam Repub Iran. 2016;30:428. [PubMed ID: 28210593]. [PubMed Central ID: PMC5307619].
-
30.
Zahedi Bialvaei A, Pourlak T, Aghamali M, Asgharzadeh M, Gholizadeh P, Kafil HS. The Prevalence of CTX-M-15 Extended-spectrum beta-Lactamases Among Salmonella spp. and Shigella spp. Isolated from three Iranian Hospitals. Eur J Microbiol Immunol (Bp). 2017;7(2):133-7. [PubMed ID: 28690880]. [PubMed Central ID: PMC5495085]. https://doi.org/10.1556/1886.2017.00004.
-
31.
Esmaeili Dooki MR, Rajabnia R, Barari Sawadkohi R, Mosaiebnia Gatabi Z, Poornasrollah M, Mirzapour M. Bacterial entropathogens and antimicrobial susceptibility in children with acute diarrhea in Babol, Iran. Caspian J Intern Med. 2014;5(1):30-4. [PubMed ID: 24490011]. [PubMed Central ID: PMC3894468].
-
32.
Ranjbar R, Mirsaeed Ghazi F. Antibiotic sensitivity patterns and molecular typing of Shigella sonnei strains using ERIC-PCR. Iran J Public Health. 2013;42(10):1151.
-
33.
Ranjbar R, Memariani M. Multilocus variable-number tandem-repeat analysis for genotyping of Shigella sonnei strains isolated from pediatric patients. Gastroenterol Hepatol Bed Bench. 2015;8(3):225-32. [PubMed ID: 26328045]. [PubMed Central ID: PMC4553163].
-
34.
Dibaj R, Aslani MM, Heidarieh P, Hashemi A, Ataei B, Daei-Naser A, et al. Conventional and molecular investigation of Shigella isolates in relation to an outbreak in the area of Isfahan, Iran. Iran J Microbiol. 2013;5(4):339-44. [PubMed ID: 25848502]. [PubMed Central ID: PMC4385158].
-
35.
Soltan Dallal MM, Ghahremani R, Akhavan SA, Rajabi Z. Frequency, Antimicrobial Resistance and Serotyping of Shigella-Contaminted Food Samples in Foodborne Disease Outbreaks in Iran. SJSPH. 2019;17(2):110-20.
-
36.
Farshad S, Ranjbar R, Hosseini M. Molecular Genotyping of Shigella sonnei Strains Isolated From Children With Bloody Diarrhea Using Pulsed Field Gel Electrophoresis on the Total Genome and PCR-RFLP of IpaH and IpaBCD Genes. Jundishapur J Microbiol. 2015;8(1). e14004. [PubMed ID: 25789126]. [PubMed Central ID: PMC4350046]. https://doi.org/10.5812/jjm.14004.
-
37.
Samie A, Guerrant RL, Barrett L, Bessong PO, Igumbor EO, Obi CL. Prevalence of intestinal parasitic and bacterial pathogens in diarrhoeal and non-diarroeal human stools from Vhembe district, South Africa. J Health Popul Nutr. 2009;27(6):739-45. [PubMed ID: 20099757]. [PubMed Central ID: PMC2928113]. https://doi.org/10.3329/jhpn.v27i6.4325.
-
38.
Cunningham SA, Sloan LM, Nyre LM, Vetter EA, Mandrekar J, Patel R. Three-hour molecular detection of Campylobacter, Salmonella, Yersinia, and Shigella species in feces with accuracy as high as that of culture. J Clin Microbiol. 2010;48(8):2929-33. [PubMed ID: 20519461]. [PubMed Central ID: PMC2916566]. https://doi.org/10.1128/JCM.00339-10.
-
39.
Sur D, Ramamurthy T, Deen J, Bhattacharya SK. Shigellosis : challenges & management issues. Indian J Med Res. 2004;120(5):454-62. [PubMed ID: 15591629].
-
40.
Muthuirulandi Sethuvel DP, Devanga Ragupathi NK, Anandan S, Veeraraghavan B. Update on: Shigella new serogroups/serotypes and their antimicrobial resistance. Lett Appl Microbiol. 2017;64(1):8-18. [PubMed ID: 27783408]. https://doi.org/10.1111/lam.12690.
-
41.
Gupta A, Polyak CS, Bishop RD, Sobel J, Mintz ED. Laboratory-confirmed shigellosis in the United States, 1989-2002: epidemiologic trends and patterns. Clin Infect Dis. 2004;38(10):1372-7. [PubMed ID: 15156473]. https://doi.org/10.1086/386326.
-
42.
von Seidlein L, Kim DR, Ali M, Lee H, Wang X, Thiem VD, et al. A multicentre study of Shigella diarrhoea in six Asian countries: disease burden, clinical manifestations, and microbiology. PLoS Med. 2006;3(9). e353. [PubMed ID: 16968124]. [PubMed Central ID: PMC1564174]. https://doi.org/10.1371/journal.pmed.0030353.
-
43.
Chompook P, Samosornsuk S, von Seidlein L, Jitsanguansuk S, Sirima N, Sudjai S, et al. Estimating the burden of shigellosis in Thailand: 36-month population-based surveillance study. Bull World Health Organ. 2005;83(10):739-46. [PubMed ID: 16283050]. [PubMed Central ID: PMC2626425].
-
44.
CDC. National Antimicrobial Resistance Monitoring System for Enteric Bacteria (NARMS): Human Isolates Surveillance Report for 2015 (Final Report). Atlanta, Georgia: U.S. Department of Health and Human Services, CDC; 2018. Available from: http://wwwn.cdc.gov/narmsnow.