Abstract
Background:
Candida albicans is one of the most important members of the human normal flora that can cause opportunistic fungal infections. Hydrolytic enzymes are one of the main virulence factors in the pathogenesis of Candida species.Objectives:
This study was carried out to determine proteolytic activities, and their related gene expressions in C. albicans isolates obtained from oropharyngeal candidiasis in head and neck cancer patients.Methods:
Thirty-two C. albicans clinical isolates were included in this study. Secreted aspartyl protease and phospholipase activities were analyzed by appropriate agar media and precipitation zones. The expression levels of SAP1, 3 and PLB1, 2 genes were evaluated by real-time PCR.Results:
All the 32 isolates exhibited proteinase activity while 28 of them showed phospholipase activity. All the strains possessed all SAPs genes; however, PLBs genes were not expressed in four isolates.Conclusions:
Our findings demonstrated that the clinical strains of C. albicans had strong proteolytic activity and high expression levels of the pertaining genes.Keywords
Candida albicans Aspartyl Proteinase Phospholipase Gene Expression
1. Background
Candida is a pathogenic fungal genus in humans whose members are responsible for 34% of the opportunistic fungal infections (1). Candida albicans is one of the most important known human pathogens that can take various clinical forms (2-4). Numerous virulence factors have been revealed in C. albicans, among which, secreted aspartyl proteinases (Saps) and phospholipases are known as extracellular hydrolytic enzymes in Candida species (5). Saps are considered as the most significant factors in the virulence of C. albicans encoded by 10 SAPs (6, 7). It has been demonstrated that SAP1 - 8 gene products are released into the extracellular spaces while the products of SAP9 and SAP10 genes are attached to the cell membrane (8). Moreover, research has revealed that SAP gene expressions are related to environmental conditions (9).
It has been found that SAP1 and SAP3 are critical for mucosal infections. Moreover, they are involved in yeast sticking, cell harm, and escape from the host immune system (9). The SAP1 and SAP3 genes are known to have significant functions in oral candidiasis and contribute to tissue damage in C. albicans infections at mucosal surfaces and SAP2 is highly expressed in oral candidiasis (10, 11). Altogether, the Sap enzymes have important roles in the colonization and invasion of Candida species (10). Among four reported types of phospholipases in C. albicans, phospholipase Plb1 and Plb2 are produced extracellularly (9) while Plb1 has been demonstrated to be a major virulence factor (12). Most phospholipase B activities in C. albicans belong to Plb1, whereas Plb2 has a slight activity; hence, the strains without the PLB1 gene only have a little phospholipase B activity (13).
Oropharyngeal candidiasis is a common oral complication that occurs after radiotherapy in head and neck cancer patients. The infection may cause critical and systemic infections (14). The literature shows that C. albicans is one of the significant species involved in more than 80% of oropharyngeal candidiasis cases (15). In our previous study, we examined SAP2 gene expression in C. albicans strains, isolated from oropharyngeal candidiasis patients with head and neck cancer in Iran (16). Due to the importance of extracellular Candida proteinases for the improvement of current therapeutic strategies and their considerable potential as fungal drug targets (17), here we evaluated the activities of these enzymes and expressions of SAP1, SAP3, PLB1, and PLB2 genes in C. albicans strains. To the best of our knowledge, this is the first report on the expression of the above-mentioned genes concerning this mucosal infection in Iran, which may improve our knowledge of the virulence factors of C. albicans in oropharyngeal candidiasis of head and neck cancer patients.
2. Objectives
In this study, we surveyed the proteolytic activity of clinical isolates of C. albicans with an emphasis on the expression of the related genes.
3. Methods
3.1. Clinical Isolates
Thirty-two clinical C. albicans isolates, as the causative agents of oropharyngeal candidiasis (typed by MLST), were obtained from patients in six geographical regions of Iran and used in this study (16). Five C. albicans isolates from head and neck cancer patients who did not have oropharyngeal candidiasis were considered as the control. The isolates were recognized using culture on CHROMagar Candida, germ tube production test, chlamydoconidia production, and carbohydrate assimilation test using the API kit (BioMerieux, France). The final identification was done by the ITS sequencing method. The primers of internal transcribed spacer regions (ITS 1 and ITS 4) were 5’- TCCGTAGGTGAACCTGCGG-3’ (forward) and 5’-TCCTCCGCTTATTGATATGC-3’ (reverse). The new sequences were submitted to the GenBank database and gave MW009706-MW009737 accession numbers for 32 sequences.
3.2. Chemicals and Instruments
Sabouraud dextrose agar (SDA), CHROMagar medium, and bovine serum albumin (BSA) agar medium were obtained from Merck (E. Merck, Germany). Guanidium isothiocyanate (GITC) reagent, RNase-free DNase, random hexamer primers, Revert Aid M-MuLV reverse transcriptase, and SYBR Green master mix were obtained from Thermo Fisher Scientific (USA). The API kit was obtained from BioMerieux (France). Agarose powder was obtained from Yekta Tajhiz Azma (YTA, Iran). An EPOCH2 microplate reader was purchased from BioTek instruments (BioTek, USA). Corbett Rotor-Gene 6000 was purchased from QIAGEN (USA).
3.3. Proteolytic Activity
The proteolytic activity of Candida isolates was investigated by bovine serum albumin (BSA) agar medium as previously specified by Ruchel et al. (18). Candida isolates were cultured as spots and were incubated at 37°C for five days. Each isolate was cultured three times. After incubation, the enzymatic activity was estimated as the diameter of the whitening halo surrounding each colony resulting from protein degradation. The ratio of the diameter of a colony to the diameter of that colony plus the precipitation zone (i.e. the total diameter) indicated the proteinase zone (Prz). The Prz values were categorized as follows: Prz value =1 (no activity), Prz value = 0.64 to 1 (moderate activity), and Prz value < 0.64 (high activity) (19).
3.4. Phospholipase Activity
For the determination of phospholipase activity of the strains, culturing on egg yolk agar medium was done (19). The culture medium consisted of SDA in which 57.3 g NaCl, 0.55 g CaCl2, and 8% sterile egg yolk emulsion were added. The effects of extracellular phospholipase enzymes on the egg yolk were characterized as phospholipase zone (Pz). Candida isolates were inoculated in triplicate. After incubation at 37°C, the diameters of the colonies and the diameters of the colonies plus precipitation zone around the colonies were measured, as well as the Pz, were calculated after 3 - 8 days. The average Pz was measured for all the isolates and phospholipase activity was measured using Pz values as mentioned above.
3.5. RNA Extraction
After culturing, the isolates on the SDA medium were incubated at 28°C for 48 h. Fungal cells were homogenized by glass beads. Total RNA was extracted using GITC reagent from fresh yeast cells (16, 20). Total RNA was treated by RNase-free DNase (Thermo Fisher Scientific, USA) according to the manufacturer’s protocol. Agarose gel electrophoresis was performed to confirm the absence of DNA. The concentration of RNA was measured by an EPOCH2 microplate reader (Bio Tek, USA). A total of 1000 ng RNA was used for preparing single-stranded cDNA (Thermo Fisher Scientific, USA) according to the manufacturer’s instructions.
3.6. SAPs and PLBs Gene Expression by Real-Time PCR (RT-PCR)
Genes expressions were evaluated by RT-PCR (Rotor gene 6000, Corbett) using the SYBR green master mix kit (Applied Biosystems). Each reaction was prepared in a final volume of 25 µL using the specific primers set, as shown in Table 1 (9). The optimized program of PCR was as follows: 95°C for 10 min, 40 cycles of 95°C for 15 s, and 60°C for 1 min (9). Each reaction was repeated three times. The results were analyzed by relative quantification, using ACT1 expression as the reference gene. Then, ΔCT was determined by the following formula: [ΔCT = CT(target) - CT (reference)]. The gene expression level was calculated by the 2-ΔCT method. Fold increases (FI) were determined using the relative threshold method (2-ΔΔCT) (16).
The Primers set Used in Real-Time PCR, SAPs: Secreted Aspartyl Proteinase, PLBs: Phospholipase B, Act 1: β-Actin
| Gene | Primers | Sequences (5’-3’) | Size of Amplified Product, bp |
|---|---|---|---|
| SAP1 | SAP1F | TCAATCAATTTACTCTTCCATTTCTAACA | 161 |
| SAP1R | CCAGTAGCATTAACAGGAGTTTTAATGACA | ||
| SAP3 | SAP3F | CCTTCTCTAAAATTATGGATTGGAAC | 231 |
| SAP3R | TTGATTTCACCTTGGGGACCAGTAACATTT | ||
| PLB1 | PLB1F | CCTATTGCCAAACAAGCATTGTC | 181 |
| PLB1R | CCAAGCTACTGATTTCACCTGCTCC | ||
| PLB2 | PLB2F | GTGGGATCTTGCAGAGTTCAAGC | 179 |
| PLB2R | CTCAAAGCTCTCCCATAGACATCTG | ||
| ACT1 | ACT1F | GACCGAAGCTCCAATGAATC | 270 |
| ACT1R | AATTGGGACAACGTGGGTAA |
3.7. Statistical Analysis
The data were analyzed using GraphPad PRISM 6 by One-Way ANOVA. A P-value of less than 0.05 (P < 0.05) was considered significant.
4. Results
4.1. Proteinase and Phospholipase Activity of Candida Isolates
All the strains exhibited proteinase activity. Twenty-eight strains (87.5%) showed a potent proteinase activity (++) and four strains (12.5%) had a moderate activity (+). Out of 32 isolates of C. albicans, 17 (53.1%) isolates revealed a strong phospholipase activity, 11 (34.3%) isolates showed a moderate phospholipase activity, and four (12.5 %) isolates had no phospholipase activity (Table 2).
The Expression of SAPs and PLBs Genes in 32 Candida albicans Isolates
| Gene Expression | |
|---|---|
| Genotypes | No. (%) |
| SAPs | |
| SAP1/SAP3 | 17 (53.12) |
| SAP1 | 11 (34.37) |
| SAP3 | 3 (9.37) |
| - | 1 (3.12) |
| PLBs | |
| PLB1/PLB2 | 24 (75) |
| PLB1 | 3 (9.37) |
| PLB2 | 1 (3.125) |
| - | 4 (12.5) |
4.2. SAPs and PLBs Gene Expression by RT- PCR
The expressions of SAP1, SAP3, PLB1, and PLB2 genes in all Candida isolates were studied by RT-PCR and compared to C. albicans SC5314 as a standard isolate (21). Candida albicans SC5314 is a pathogenic strain widely used for genomic and experimental studies. Invasive hyphal forms are observed in this strain, and its genome is available at the Candida genome database (CGD) (22, 23). The SAP1 and SAP3 genes were expressed by most C. albicans strains isolated from head and neck cancer patients with oropharyngeal candidiasis. The SAP1 gene was expressed in 28 isolates, while SAP3 was expressed in 20 isolates (Figure 1). The PLB1 gene was expressed in most isolates (Figure 2). The RT-PCR amplifications revealed that all the isolates expressed the SAP genes; however, the PLB genes were expressed in none of them.
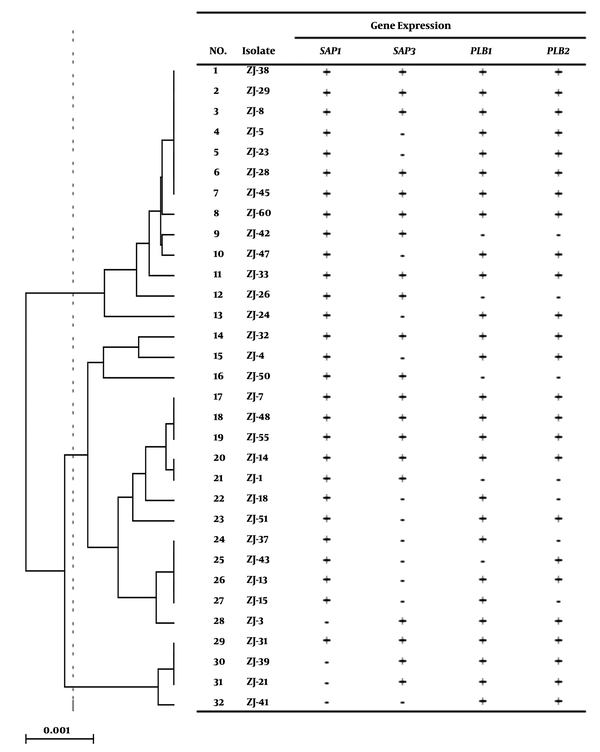
As shown in Table 2, 17 (53.12%) isolates expressed the SAP1 and SAP3 genes, 11 (34.37%) isolates SAP1, three (9.37%) isolates SAP3, and one (2.86%) isolate did not express the SAP genes. The expression of PLBs genes in the isolates had variations. Twenty-four (75%) isolates showed the expressions of PLB1 and PLB2 while four (12.5%) isolates had no PLBs gene expression (Table 2). According to Figure 3, gene expressions were different in three major clades of MLST (16). Candida albicans strains of two major MLST clades expressed the SAP1 gene while the strains of the first clade showed a higher expression. Among C. albicans strains of these clades, only one strain did not express SAP1. In the second clade, SAP3 had the highest expression. Meanwhile, PLB1 and PLB2 were highly expressed in the two first clades; therefore, many strains did not have PLB2 expression.
The expression of SAP1 and SAP3 genes in 32 Candida albicans isolates and five isolates from non-oropharyngeal candidiasis patients as controls (C1 to C5). β-actin was considered as a reference gene. The mean ± SD values are shown on each bar.
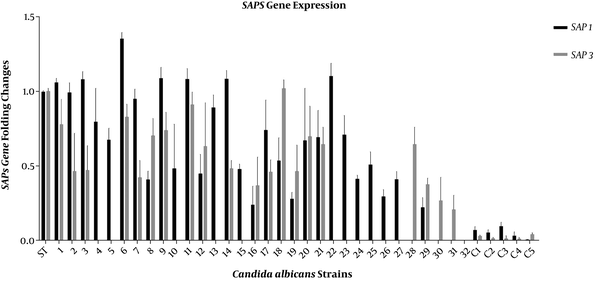
The expression of PLB1 and PLB2 genes in 32 Candida albicans isolates and five isolates from non-oropharyngeal candidiasis patients as controls (C1 to C5). β-actin was considered as a reference gene. The mean ± SD values are shown on each bar.
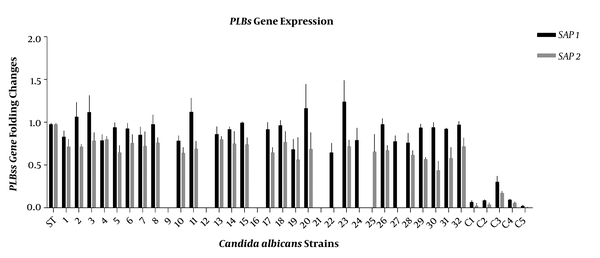
Gene expression pattern of 32 Candida albicans strains on UPGMA dendrogram of seven MLST loci of C. albicans strains
5. Discussion
Candida albicans is a frequent cause of Candida infections in patients with cancer and oropharyngeal candidiasis and is also one of the most frequent causes of oral problems in head and neck cancer patients. It has been demonstrated that there is a significant relationship between pathogenicity and proteinase activity in Candida spp. Moreover, the duration of C. albicans infection is related to the total number of virulence factors (24, 25). The present study investigated the secreted aspartyl proteinase and phospholipase activities of 32 C. albicans strains obtained from oropharyngeal candidiasis lesions of head and neck cancer patients, as well as the expression of SAP1, SAP3, PLB1, and PLB2 genes in the isolates.
We had previously shown by RT-PCR analysis that the same 32 C. albicans isolates had high expression levels of the SAP2 gene (16). Our present results showed that the SAP1 and SAP3 genes were expressed in more than half of the C. albicans isolates. Although SAP2 is most commonly expressed during oral C. albicans infections, it has been found that the SAP1 and SAP3 genes are preferentially expressed and have principle roles in oral candidiasis (10, 11). In the present study, we also showed that SAP1 had a higher level of expression than SAP3, as SAP1 was expressed in 87.5% (n = 28) and SAP3 was expressed in 62.5% (n = 20) of the oral clinical isolates of C. albicans. As shown in Figure 1, the expression of SAP genes was much lower in the control group than in the test group. When the isolates were categorized into four groups based on their SAP1 and SAP3 expressions (Table 2), most of them belonged to the group in which both SAP genes were expressed (53.12%, n = 17).
It has been demonstrated that C. albicans strains with increased SAP2 expression appear to be contributing to severe infections such as HIV (26). Studies have also indicated that SAP2 gene expression is higher than other SAP gene expressions in samples isolated from oral infections (7). Santosh et al. observed significant differences in the ability of aspartyl proteinase enzyme production between non-albicans strains (0.17 - 0.61 µg/mL) and C. albicans strains (0.56 to 0.82 µg/mL); thus, C. albicans strains were more potent secretors of aspartyl proteinase enzymes than non-albicans strains (27). It has been demonstrated that phospholipase and proteinase activities of C. albicans are considered essential virulence factors and lack of them or their decreased activities may contribute to the reduced virulence of some Candida species (28). Studies have shown that during the course of candidiasis, all the SAP genes would be expressed by C. albicans isolates in vivo; however, there are differential expressions of specific hydrolytic enzymes during oral infections (9). These findings are well in agreement with our data.
It has also been found that SAP1 and SAP3 expressions are closely coupled and are related to phenotypic switching (29, 30); and they are expressed during oral and vaginal candidiasis; however, their expressions are more common during oral infections (9). Although the expression of the SAP3 gene is higher in active mucosal infections, it is not expressed in all kinds of mucosal infections. The SAP3 expression is more common in patients with vaginal infections or carriers (9). It has been demonstrated that individual SAP and PLB genes are more commonly expressed during active infections of C. albicans (9). Studies have revealed that PLB1 regulates the gene expression of the hyphal phase (31) and is expressed during human oral infections. Moreover, it has been reported that PLB1 is expressed more than PLB2 during oral and vaginal infections (13). Here, we showed that all the isolates, except four of them, had phospholipase activity.
The results of RT-PCR confirmed four isolates with no PLB (PLB1 and PLB2) gene expression. We also found that PLB1 expression was more than PLB2 expression in C. albicans isolates and similar to previous studies, no correlation was found between the expression of SAP and PLB genes (9, 13). Our results also revealed that 22 (75%) isolates expressed both PLB1 and PLB2 genes while the mean mRNA level of PLB1 was higher than that of PLB2. The control group expressed lower levels of PLBs genes than the test group (Figure 2). We had previously evaluated the production of secretory enzymes, i.e. Saps and phospholipase, and their encoding genes in C. albicans strains obtained from oropharyngeal candidiasis in head and neck cancer patients and had compared the three major clades derived from the MLST typing method (16). It has been demonstrated that Candida strains from oral or systemic infections can be used to determine the genetic factors influencing the Candida virulence ability (32). Here, all C. albicans isolates of the three major clades represented different levels of the studied virulence factors, although we could not find a significant relationship between the virulence factors and MLST clades.
It has been found that small modifications in the sequence of specific genes can change phenotypic specifications of C. albicans over a long period (33). Tavanti et al. (34) have reported that the utility of MLST for determining clade assignments of clinical isolates provides a basis for the rational selection of more extensive diversity of the strains for future studies on C. albicans virulence factors. It seems that the fungal species and the host immunity have important roles in the determination of the fungal infection severity and extension of clinical appearance (34). It was reported that the relationship between the virulence factors and MLST clades in C. albicans was significant when samples with different infectious sources and various geological regions were used in the study (35). Here, some isolates had no expression of a specific gene although we could not find important correlations between the expressed genes and MLST genotype clades in C. albicans strains.
5.1. Conclusions
In conclusion, we showed the importance of aspartyl proteinase and phospholipase activities and their contributions to the virulence and pathogenesis of C. albicans with an emphasis on their gene expressions in oral infection of head and neck cancer patients. Our results revealed that most C. albicans isolates could produce protease and phospholipase, which are crucial enzymes in the infection process of C. albicans. We also found considerable strain-to-strain variations concerning the produced enzymes in these patients. The expression of SAPs and PLBs genes may be correlated with the severity of the infection. Further investigations are needed to elucidate the role of proteolytic enzymes and their protease activity in C. albicans.
References
-
1.
Pfaller MA, Diekema DJ. Epidemiology of invasive candidiasis: a persistent public health problem. Clin Microbiol Rev. 2007;20(1):133-63. [PubMed ID: 17223626]. [PubMed Central ID: PMC1797637]. https://doi.org/10.1128/CMR.00029-06.
-
2.
Minooeianhaghighi MH, Sehatpour M, Zarrinfar H, Sen T. Recurrent vulvovaginal candidiasis: the causative agents, clinical signs and susceptibility to fluconazole in Gonabad city. Curr Women Health Rev. 2020;16(1):46-51. https://doi.org/10.2174/1573404815666191104142813.
-
3.
Zarrinfar H, Kaboli S, Dolatabadi S, Mohammadi R. Rapid detection of Candida species in bronchoalveolar lavage fluid from patients with pulmonary symptoms. Braz J Microbiol. 2016;47(1):172-6. [PubMed ID: 26887241]. [PubMed Central ID: PMC4822774]. https://doi.org/10.1016/j.bjm.2015.02.001.
-
4.
Kord Z, Fata A, Zarrinfar H. Molecular Identification of Candida species isolated from patients with vulvovaginitis for the first time in Mashhad. Iran J Obstet Gynecol Infertil. 2017;20(4):50-7. https://doi.org/10.22038/IJOGI.2017.8982.
-
5.
Naglik JR, Challacombe SJ, Hube B. Candida albicans secreted aspartyl proteinases in virulence and pathogenesis. Microbiol Mol Biol Rev. 2003;67(3):400-28. table of contents. [PubMed ID: 12966142]. [PubMed Central ID: PMC193873]. https://doi.org/10.1128/mmbr.67.3.400-428.2003.
-
6.
Kumar CP, Kumar SS, Menon T. Phospholipase and proteinase activities of clinical isolates of Candida from immunocompromised patients. Mycopathologia. 2006;161(4):213-8. [PubMed ID: 16552483]. https://doi.org/10.1007/s11046-005-0157-4.
-
7.
Kuriyama T, Williams DW, Lewis MA. In vitro secreted aspartyl proteinase activity of Candida albicans isolated from oral diseases and healthy oral cavities. Oral Microbiol Immunol. 2003;18(6):405-7. [PubMed ID: 14622349]. https://doi.org/10.1046/j.0902-0055.2002.00100.x.
-
8.
Modrzewska B, Kurnatowski P, Khalid K. Comparison of proteolytic activity of Candida sp. strains depending on their origin. J Mycol Med. 2016;26(2):138-47. [PubMed ID: 26922385]. https://doi.org/10.1016/j.mycmed.2016.01.005.
-
9.
Naglik JR, Rodgers CA, Shirlaw PJ, Dobbie JL, Fernandes-Naglik LL, Greenspan D, et al. Differential expression of Candida albicans secreted aspartyl proteinase and phospholipase B genes in humans correlates with active oral and vaginal infections. J Infect Dis. 2003;188(3):469-79. [PubMed ID: 12870130]. https://doi.org/10.1086/376536.
-
10.
Silva NC, Nery JM, Dias AL. Aspartic proteinases of Candida spp.: role in pathogenicity and antifungal resistance. Mycoses. 2014;57(1):1-11. [PubMed ID: 23735296]. https://doi.org/10.1111/myc.12095.
-
11.
Rossetti B, Lombardi F, Belmonti S, D'Andrea MM, Tordini G, D'Avino A, et al. Nasopharingeal bacterial and fungal colonization in HIV-positive versus HIV-negative adults. New Microbiol. 2019;42(1):37-42. [PubMed ID: 30671585].
-
12.
Samaranayake YH, Dassanayake RS, Cheung BP, Jayatilake JA, Yeung KW, Yau JY, et al. Differential phospholipase gene expression by Candida albicans in artificial media and cultured human oral epithelium. APMIS. 2006;114(12):857-66. [PubMed ID: 17207086]. https://doi.org/10.1111/j.1600-0463.2006.apm_479.x.
-
13.
Ghannoum MA. Potential role of phospholipases in virulence and fungal pathogenesis. Clin Microbiol Rev. 2000;13(1):122-43. table of contents. [PubMed ID: 10627494]. [PubMed Central ID: PMC88936]. https://doi.org/10.1128/cmr.13.1.122-143.2000.
-
14.
Suryawanshi H, Ganvir SM, Hazarey VK, Wanjare VS. Oropharyngeal candidosis relative frequency in radiotherapy patient for head and neck cancer. J Oral Maxillofac Pathol. 2012;16(1):31-7. [PubMed ID: 22438640]. [PubMed Central ID: PMC3303519]. https://doi.org/10.4103/0973-029X.92970.
-
15.
Tati S, Davidow P, McCall A, Hwang-Wong E, Rojas IG, Cormack B, et al. Candida glabrata Binding to Candida albicans Hyphae Enables Its Development in Oropharyngeal Candidiasis. PLoS Pathog. 2016;12(3). e1005522. [PubMed ID: 27029023]. [PubMed Central ID: PMC4814137]. https://doi.org/10.1371/journal.ppat.1005522.
-
16.
Jahanshiri Z, Manifar S, Hatami F, Arastehnazar F, Shams-Ghahfarokhi M, Razzaghi-Abyaneh M. Genotyping of Candida albicans isolates from oropharyngeal candidiasis in head and neck cancer patients in Iran: Molecular epidemiology and SAP2 gene expression. J Mycol Med. 2019;29(4):310-6. [PubMed ID: 31630914]. https://doi.org/10.1016/j.mycmed.2019.100896.
-
17.
Rapala-Kozik M, Bochenska O, Zajac D, Karkowska-Kuleta J, Gogol M, Zawrotniak M, et al. Extracellular proteinases of Candida species pathogenic yeasts. Mol Oral Microbiol. 2018;33(2):113-24. [PubMed ID: 29139623]. https://doi.org/10.1111/omi.12206.
-
18.
Ruchel R, Tegeler R, Trost M. A comparison of secretory proteinases from different strains of Candida albicans. Sabouraudia. 1982;20(3):233-44. [PubMed ID: 6753190]. https://doi.org/10.1080/00362178285380341.
-
19.
Price MF, Wilkinson ID, Gentry LO. Plate method for detection of phospholipase activity in Candida albicans. Sabouraudia. 1982;20(1):7-14. [PubMed ID: 7038928]. https://doi.org/10.1080/00362178285380031.
-
20.
Ho HC, Shiau PF, Liu FC, Chung JG, Chen LY. Purification, characterization and complete amino acid sequence of nuclease C1 from Cunninghamella echinulata var. echinulata. Eur J Biochem. 1998;256(1):112-8. [PubMed ID: 9746353]. https://doi.org/10.1046/j.1432-1327.1998.2560112.x.
-
21.
Joo MY, Shin JH, Jang HC, Song ES, Kee SJ, Shin MG, et al. Expression of SAP5 and SAP9 in Candida albicans biofilms: comparison of bloodstream isolates with isolates from other sources. Med Mycol. 2013;51(8):892-6. [PubMed ID: 23971863]. https://doi.org/10.3109/13693786.2013.824623.
-
22.
Hua X, Yuan X, Mitchell BM, Lorenz MC, O'Day DM, Wilhelmus KR. Morphogenic and genetic differences between Candida albicans strains are associated with keratomycosis virulence. Mol Vis. 2009;15:1476-84. [PubMed ID: 19649176]. [PubMed Central ID: PMC2718853].
-
23.
Green CB, Marretta SM, Cheng G, Faddoul FF, Ehrhart EJ, Hoyer LL. RT-PCR analysis of Candida albicans ALS gene expression in a hyposalivatory rat model of oral candidiasis and in HIV-positive human patients. Med Mycol. 2006;44(2):103-11. [PubMed ID: 16519012]. [PubMed Central ID: PMC2583129]. https://doi.org/10.1080/13693780500086527.
-
24.
Bensadoun RJ, Patton LL, Lalla RV, Epstein JB. Oropharyngeal candidiasis in head and neck cancer patients treated with radiation: update 2011. Support Care Cancer. 2011;19(6):737-44. [PubMed ID: 21479787]. https://doi.org/10.1007/s00520-011-1154-4.
-
25.
Pereira CA, da Costa AC, Machado AK, Beltrame Junior M, Zollner MS, Junqueira JC, et al. Enzymatic activity, sensitivity to antifungal drugs and Baccharis dracunculifolia essential oil by Candida strains isolated from the oral cavities of breastfeeding infants and in their mothers' mouths and nipples. Mycopathologia. 2011;171(2):103-9. [PubMed ID: 20703942]. https://doi.org/10.1007/s11046-010-9353-y.
-
26.
De Bernardis F, Chiani P, Ciccozzi M, Pellegrini G, Ceddia T, D'Offizzi G, et al. Elevated aspartic proteinase secretion and experimental pathogenicity of Candida albicans isolates from oral cavities of subjects infected with human immunodeficiency virus. Infect Immun. 1996;64(2):466-71. [PubMed ID: 8550193]. [PubMed Central ID: PMC173787]. https://doi.org/10.1128/IAI.64.2.466-471.1996.
-
27.
Deorukhkar SC, Saini S, Mathew S. Virulence factors contributing to pathogenicity of Candida tropicalis and its antifungal susceptibility profile. Int J Microbiol. 2014;2014:456878. [PubMed ID: 24803934]. [PubMed Central ID: PMC3996979]. https://doi.org/10.1155/2014/456878.
-
28.
Mattei AS, Alves SH, Severo CB, Guazzelli Lda S, Oliveira Fde M, Severo LC. Determination of germ tube, phospholipase, and proteinase production by bloodstream isolates of Candida albicans. Rev Soc Bras Med Trop. 2013;46(3):340-2. [PubMed ID: 23856861]. https://doi.org/10.1590/0037-8682-0045-2013.
-
29.
White TC, Agabian N. Candida albicans secreted aspartyl proteinases: isoenzyme pattern is determined by cell type, and levels are determined by environmental factors. J Bacteriol. 1995;177(18):5215-21. [PubMed ID: 7665510]. [PubMed Central ID: PMC177311]. https://doi.org/10.1128/jb.177.18.5215-5221.1995.
-
30.
Hube B, Monod M, Schofield DA, Brown AJ, Gow NA. Expression of seven members of the gene family encoding secretory aspartyl proteinases in Candida albicans. Mol Microbiol. 1994;14(1):87-99. [PubMed ID: 7830564]. https://doi.org/10.1111/j.1365-2958.1994.tb01269.x.
-
31.
Hoover CI, Jantapour MJ, Newport G, Agabian N, Fisher SJ. Cloning and regulated expression of the Candida albicans phospholipase B (PLB1) gene. FEMS Microbiol Lett. 1998;167(2):163-9. [PubMed ID: 9809417]. https://doi.org/10.1111/j.1574-6968.1998.tb13223.x.
-
32.
Chin VK, Lee TY, Rusliza B, Chong PP. Dissecting Candida albicans infection from the perspective of C. albicans virulence and omics approaches on host-pathogen interaction: a review. Int J Mol Sci. 2016;17(10). [PubMed ID: 27763544]. [PubMed Central ID: PMC5085676]. https://doi.org/10.3390/ijms17101643.
-
33.
Odds FC, Jacobsen MD. Multilocus sequence typing of pathogenic Candida species. Eukaryot Cell. 2008;7(7):1075-84. [PubMed ID: 18456859]. [PubMed Central ID: PMC2446668]. https://doi.org/10.1128/EC.00062-08.
-
34.
Tavanti A, Davidson AD, Fordyce MJ, Gow NA, Maiden MC, Odds FC. Population structure and properties of Candida albicans, as determined by multilocus sequence typing. J Clin Microbiol. 2005;43(11):5601-13. [PubMed ID: 16272493]. [PubMed Central ID: PMC1287804]. https://doi.org/10.1128/JCM.43.11.5601-5613.2005.
-
35.
MacCallum DM, Castillo L, Nather K, Munro CA, Brown AJ, Gow NA, et al. Property differences among the four major Candida albicans strain clades. Eukaryot Cell. 2009;8(3):373-87. [PubMed ID: 19151328]. [PubMed Central ID: PMC2653250]. https://doi.org/10.1128/EC.00387-08.