Abstract
Background:
Non-tuberculous mycobacteria (NTM) are widely associated with pulmonary diseases. Evidence is lacking on the transmission of NTM from one person to another. Hence, it has gained lower public health priority than tuberculosis.Objectives:
We determined the prevalence and antibiotic resistance rate of NTM isolated from sputum samples of patients with pulmonary infections.Methods:
A total of 375 duplicate sputum samples were collected from 375 patients on consecutive days. The NTM growth was assessed using BACTEC 960 mycobacterial growth indicator tubes. The GenoType Mycobacterium CM/AS line probe assay was used for the species-level identification of mycobacteria. Antibiotic susceptibility tests were performed using the auto-MODS assay.Results:
The overall NTM prevalence rate was 34.4%. Mycobacterium avium complex (24.8%) was the predominant species identified, followed by M. kansasii (24%) and M. abscessus complex (20.2%). Of the 129 NTM isolates tested for antibiotic susceptibility, 62.8% were resistant to rifampicin, 60.5% to levofloxacin, 58.1% to ofloxacin, 55.8% to ethambutol, 49.6% to isoniazid, 48.1% to streptomycin, and 41.9% to amikacin. Seventy-three (56.6%) isolates were identified as multidrug-resistant (MDR) isolates.Conclusions:
Mycobacterium avium complex was the predominant species identified, and the majority of the organisms were resistant to commonly used anti-tuberculosis drugs. The high prevalence of NTM and drug resistance towards the tested antibiotics suggests that NTM can no more be ignored as a contaminant, reiterating the need for periodic surveillance and species-specific treatment for effective management of diseases caused by NTM.Keywords
Non-tuberculous Mycobacteria Auto-MODS Antimicrobial Susceptibility
1. Background
The non-tuberculous mycobacteria (NTM) represent the Mycobacterium species that are not members of the Mycobacterium leprae and M. tuberculosis complex (1). Non-tuberculous mycobacteria are ubiquitous mycobacterial species that are found worldwide and cause various infections, particularly pulmonary infections that are difficult to diagnose and treat (2). They have also been widely associated with pulmonary diseases, of which chronic pulmonary disease is the most common clinically encountered syndrome (3). Due to the lack of definitive evidence on the transmission of NTM from one person to another, NTM has gained lower public health priority than tuberculosis (4). The incidence and prevalence of NTM lung diseases are increasing worldwide. The common causative organisms of pulmonary infections include slow-growing mycobacteria (SGM), including M. avium complex (MAC) and M. kansasii, and rapid-growing mycobacteria (RGM) including M. abscessus complex (5).
The percentage of NTM lung infection increases with age but varies significantly between countries (6). The prevalence and the mortality rate due to NTM disease have been progressively rising worldwide (2). A study conducted between 2004 and 2006 in the USA suggested that the mean annual site-specific prevalence rates ranged from 1.4 to 6.6 per 100,000 individuals (7). Another population-based study conducted in five states of the USA revealed that the estimated NTM disease prevalence increased from 9.2 to 15.2 per 100,000 persons between the years 2008 and 2013 (8). A study from the UK reported an increase in the incidence of pulmonary NTM-positive cultures from 4.0 to 6.1 per 100,000 people from 2007 to 2012 (9). Another study revealed that the annual prevalence of NTM in a population with > 65-years-old significantly increased from 20 to 47 cases/100,000 persons from 1997 to 2007. Of these, 90% of the individuals were Caucasians, and the remaining persons were Blacks and Asians/Pacific Islanders (6). In China, the isolation rate of NTM increased from 3% in 2008 to 8.5% in 2012 (10). Non-tuberculous mycobacteria usually cause chronic lung infection and are recognized as one of the major emerging pathogens. They are often misdiagnosed as multidrug-resistant tuberculosis (MDR TB) due to the difficulty in distinguishing the clinical symptoms between TB and NTM disease (11).
Non-tuberculous mycobacteria are also resistant to many of the first- and second-line drugs used to treat tuberculosis; hence, an accurate diagnosis and drug-susceptibility testing are crucial for effective clinical management of NTM infections (12). A study from China reported high drug resistance by NTM to first-line anti-TB drugs, of which 30.7% of the isolates were suspected to be of MDR-TB cases and 4.0% of TB re-treatment cases. The results indicate that pulmonary infections caused by NTM may lead to substantial difficulties in the treatment of NTM and MDR-TB diseases in China (13). The microscopic observation drug susceptibility (MODS) assay is an economical, highly sensitive, and rapid method for the detection of M. tuberculosis (14). The World Health Organization (WHO) has recommended MODS as an affordable and highly effective alternative to existing gold standard liquid mycobacterial culture methods for testing sputum samples of TB suspected individuals (15). Although MODS has several advantages, due to concerns about the biosafety and efficiency of large sample handling, the use of MODS is very limited in resource-limited settings (16).
In high-TB-burdened settings, a large number of sputum samples are to be processed, and doing MODS requires more resources and time due to the manual reading of every well by trained laboratory professionals. Furthermore, the ability of MODS in differentiating M. tuberculosis and NTM is questionable (17). To overcome these shortcomings, a recent study by Wang et al. (16) attempted to evaluate the automated MODS (auto-MODS) assay and evidenced that in resource-limited settings, auto-MODS could be a cost-sensitive and effective alternate diagnostic tool for TB diagnosis (16).
2. Objectives
The present study determined the prevalence and antimicrobial susceptibility of non-tuberculous mycobacteria isolated from sputum samples of patients with pulmonary infections.
3. Methods
3.1. Patients and Samples
Consecutive patients who were suspected of pulmonary and extra-pulmonary tuberculosis and admitted to the Department of Laboratory Medicine, Danyang People’s Hospital of Jiangsu Province, China, between April 2018 and September 2019 were included. Patients with confirmed TB were excluded from the study. Informed consent was obtained from each patient or their legal heirs. Early morning well-coughed sputum samples were collected. The collected sputum samples were stored immediately at 2 - 4°C and processed within seven days. Sputum samples were processed using modified Petroff’s method, as described by Tripathi et al. (18). Briefly, 3 - 5 mL of the sputum sample was homogenized with an equal volume of 4% NaOH in a shaker for 15 min. The homogenate was spinning for 15 min at 3,000 rpm. Then, the supernatant was discarded, and 20 mL of sterile distilled water was added to the deposit for neutralization. Again, the sample was spinning for 15 min at 3,000 rpm. After processing, the sputum samples were split into different aliquots in a sterile screwcap container. Of these, only the acid-fast bacilli (AFB)-positive sputum samples were used for further analysis.
3.2. Bacterial Identification
The sputum sample from one screwcap vial was inoculated into BACTEC 960 mycobacterial growth indicator tubes (MGIT, BD Diagnostics, USA) and observed for 42 days. The test was performed in duplicate, and the tested sample was considered positive only if both showed growth. The GenoType Mycobacterium CM/AS line probe assay (Hain Lifescience, Nehren, Germany) was used for the species-level identification of mycobacteria.
3.3. Auto-MODS Technique
Drug susceptibility through the auto-MODS technique was performed as described by Wang et al. (17). In this study, there were four modifications proposed for the original MODS assay (14, 19). A p-nitrobenzoic acid (PNB) tube was used in the auto-MODS assay to differentiate TB and NTM. So, PNB inhibits the growth of M. tuberculosis, while NTM differentiates TB and NTM resistant to PNB (14). An automatic computer-assisted digital camera was used to reduce human resource requirement for frequent readings and the images captured were used in auto-MODS. Antibiotics tested in this study included isoniazid (0.1 µg/mL), rifampicin (1 µg/mL), ethambutol (5 µg/mL), streptomycin (1 µg/mL), amikacin (1 µg/mL), levofloxacin (2 µg/mL), and ofloxacin (2 µg/mL). Antibiotic concentrations were determined based on the GLI Mycobacteriology Laboratory Manual (20). After inoculation, the tubes were incubated at 37°C, and daily images of the tubes were taken through the computer-assisted digital camera, which was integrated with the incubator. The interpretation of auto-MODS is as same as that of the MODS assay, except that a PNB negative (no growth of TB) tube was interpreted as TB-positive, and the PNB positive tubes were interpreted as NTM-positive. The presence of growth in drug-containing tubes indicated resistance towards the drug present in the tube.
3.4. Statistical Analysis
Categorical values were represented by mean and percentages. The chi-square analysis was performed to identify significant differences. A regression analysis was performed to analyze the relationship of antimicrobial resistance with clinical features. Statistical analyses were performed using SPSS statistical software (SPSS, IMB, USA).
4. Results
A total of 375 sputum samples (duplicate samples collected on consecutive days) were collected from 375 patients. Of the 375 patient samples, 219 samples were positive in AFB staining. Thirty-two AFB-positive patient samples did not show any presence of growth in BACTEC 960. Hence, these patients were excluded from the analysis. One hundred and eighty-seven patient samples, which were detected with either NTM or TB, were included in this analysis. Of the 187 patient samples, NTM was identified in 129 (69%) patient samples and TB in 58 (31%) patient samples. Of the 187 patients (mean age 49.7 ± 7.2 years; age range 26 to 84 years), 113 (60.4%) were males, and 74 (39.6%) were females. The majority of the included patients (n = 74, 39.6%) were in the age group of 51 - 70 years, followed by 61 (32.6%) patients in the age group of 31 - 50 years.
One hundred and four (55.6%) patients had a history of smoking, and 35 (18.7%) were HIV-positive patients. Of the 129 NTM-positive patients (mean age 42.3 ± 8.1 years; age range 26 to 78 years), 79 (61.2%) were males, and 50 (38.8%) were females. The majority of the NTM-positive patients (n = 53, 41.1%) were in the age group of 51 - 70 years, followed by 39 (30.2%) patients in the age group of 31 - 50 years. Sixty-seven (51.9%) patients had a history of smoking, and 12 (9.3%) were HIV-positive patients. Except for the presence of a higher HIV rate (P = 0.0213) in the overall culture-positive patients, there was no significant difference (P > 0.05) in the demographics of the overall positive and NTM positive patients (Table 1).
Demographic Details of Overall Positive and Non-tuberculous Mycobacteria Positive Patients a
| Characteristics | Overall Positive (n = 187) | NTM Positive (n = 129) | P Values |
|---|---|---|---|
| Male | 113 (60.4) | 79 (61.2) | 0.91 |
| Female | 74 (39.6) | 50 (38.8) | 0.52 |
| Age (y), mean ± SD | 49.7 ± 7.2 | 42.3 ± 8.1 | NA |
| 21 - 30 | 21 (11.2) | 13 (10.1) | 0.07 |
| 31 - 50 | 61 (32.6) | 39 (30.2) | 0.12 |
| 51 - 70 | 74 (39.6) | 53 (41.1) | 0.09 |
| > 70 | 31 (16.6) | 24 (18.6) | 0.16 |
| Smoking history | 104 (55.6) | 67 (51.9) | 0.13 |
| HIV-positive | 35 (18.7) | 12 (9.3) | 0.02 |
| Dry cough | 121 (64.7) | 52 (40.3) | 0.07 |
Of the overall 375 patients, 129 samples were tested positive for NTM, accounting for a prevalence rate of 34.4% among our population. All the NTM-positive sputum samples were subjected to species level identification using the GenoType Mycobacterium CM/AS line probe assay. Of the 129 sputum samples, M. avium complex (n = 32, 24.8%) was the predominant species identified, followed by M. kansasii (n = 31, 24%), M. abscessus complex (n = 26, 20.2%), M. simiae (n = 21, 16.3%), and M. fortuitum (n = 11, 8.5%). No significant difference was found between the different species identified (P > 0.05). Other species were identified in fewer frequencies (Table 2).
Distribution of Non-tuberculous Mycobacteria Species Identified
| Species | No. of Isolates (%) |
|---|---|
| Mycobacterium avium complex | 32 (24.8) |
| Mycobacterium kansasii | 31 (24) |
| Mycobacterium abscessus complex | 26 (20.2) |
| Mycobacterium simiae | 21 (16.3) |
| Mycobacterium fortuitum | 11 (8.5) |
| Mycobacterium xenopi | 4 (3.1) |
| Mycobacterium gordonae | 2 (1.6) |
| Mycobacterium celonae | 2 (1.6) |
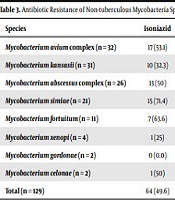
Of the 129 NTM isolates tested for antibiotic susceptibility by auto-MODS technique, 81 (62.8%) isolates were resistant to rifampicin, 78 (60.5%) to levofloxacin, 75 (58.1%) to ofloxacin, 72 (55.8%) to ethambutol, 64 (49.6%) to isoniazid, 62 (48.1%) to streptomycin, and 54 (41.9%) to amikacin. No significant difference was found in resistance between different species (P > 0.05). Of the 129 isolates tested, 73 (56.6%) isolates were identified as MDR isolates. More than 60% of the M. avium complex isolates were found to be resistant to levofloxacin (n = 22, 68.8%) and ofloxacin (n = 24, 75%). Likewise, more than 60% of M. kansasii isolates were resistant to rifampicin (n = 24, 77.4%), streptomycin (n = 21, 67.7%), and levofloxacin (n = 21, 67.7%). More than 60% of M. abscessus complex isolates were resistant to rifampicin (n = 17, 65.4%), streptomycin (n = 17, 65.4%), and ofloxacin (n = 16, 61.5%). More than 60% of M. simiae (n = 15, 71.4%) and M. fortuitum (n = 9, 81.8%) isolates were resistant to ethambutol (Table 3). The regression analysis showed a significant association between NTM isolated from HIV patients and MDR (P = 0.016). However, there was no significant association between MDR isolates and patients’ age, sex, and smoking history (P > 0.05).
| Species | Isoniazid | Rifampicin | Ethambutol | Streptomycin | Amikacin | Levofloxacin | Ofloxacin |
|---|---|---|---|---|---|---|---|
| Mycobacterium avium complex (n = 32) | 17 (53.1) | 21 (65.6) | 18 (56.3) | 12 (37.5) | 17 (53.1) | 22 (68.8) | 24 (75) |
| Mycobacterium kansasii (n = 31) | 10 (32.3) | 24 (77.4) | 12 (38.7) | 21 (67.7) | 19 (61.3) | 21 (67.7) | 17 (54.8) |
| Mycobacterium abscessus complex (n = 26) | 13 (50) | 17 (65.4) | 15 (57.7) | 17 (65.4) | 11 (42.3) | 14 (53.8) | 16 (61.5) |
| Mycobacterium simiae (n = 21) | 15 (71.4) | 11 (52.4) | 15 (71.4) | 9 (42.9) | 6 (28.6) | 12 (57.1) | 9 (42.9) |
| Mycobacterium fortuitum (n = 11) | 7 (63.6) | 3 (27.3) | 9 (81.8) | 3 (27.3) | 0 (0.0) | 3 (27.3) | 3 (27.3) |
| Mycobacterium xenopi (n = 4) | 1 (25) | 3 (75) | 2 (50) | 0 (0.0) | 0 (0.0) | 3 (75) | 3 (75) |
| Mycobacterium gordonae (n = 2) | 0 (0.0) | 1 (50) | 1 (50) | 0 (0.0) | 1 (50) | 1 (50) | 1 (50) |
| Mycobacterium celonae (n = 2) | 1 (50) | 1 (50) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 2 (100) | 2 (100) |
| Total (n = 129) | 64 (49.6) | 81 (62.8) | 72 (55.8) | 62 (48.1) | 54 (41.9) | 78 (60.5) | 75 (58.1) |
5. Discussion
The adaptive biological property and opportunistic potential of NTM make them ubiquitous microorganisms present in diverse clinical and environmental scenarios. The prevalence of NTM varies globally. They are associated with several infections, especially in immunosuppressed/immunocompromised patients (21). The association of NTM with pulmonary diseases, especially chronic pulmonary disease, is most commonly encountered in different clinical settings (3). Due to the lack of definitive evidence on the transmission of NTM from one person to another, NTM has gained lower public health priority than tuberculosis (4). Due to the existing burden of M. tuberculosis in several countries and the limited availability of rapid molecular methods, NTM is considered extremely low priority organisms (22). Besides, NTM is often misdiagnosed as MDR TB, and is being treated with anti-tuberculosis drugs such as isoniazid and rifampicin. The misdiagnosis of NTM for M. tuberculosis, the overuse of anti-tuberculosis drugs, and the selective pressure in the hospital environment have contributed to the development of resistance to several antibiotics.
The American Thoracic Society has suggested a repeat culture of sputum to establish pulmonary disease (3). Generally, NTM isolated from a single-sputum sample is considered a contaminant and often disregarded in the clinical setting. Thus, in our study, bacterial growth was assessed in duplicate samples, and a particular sputum sample was termed as NTM-positive only if both of the duplicate samples showed the presence of growth. The NTM lung infection rate increases with age but varies significantly between countries (6). In our study, the majority (41.1%) of the NTM positive patients were in the older age group of 51 - 70 years, and the rate decreased as the age group decreased. Similar to our study, a study from the United Kingdom, which reported the longitudinal trends of NTM isolation and drug susceptibility between 2000 and 2013, revealed that the majority of NTM-positive patients were in the age group of 51 - 70 years (23). In our study, HIV patients were significantly higher (P = 0.0141) in the overall AFB-positive group than in the NTM-positive group. Of the 35 HIV-positive patients, sputum samples from 19 patients showed the presence of TB. In those patients, there is a very high possibility that these patients acquired TB as a co-infection of HIV.
In the present study, the overall prevalence of NTM was 34.4% (all samples). A study from Iran reported a much lower (15.1%) prevalence than that reported in our study (24). In our study, of the 187 AFB-positive samples, NTM was identified in 129 (69%) patient samples. A study from India reported that only 3.9% of the AFB-positive samples grew NTM from various clinical samples, which is much lower than that reported in our study (21). Cowman et al. (23) reported that the proportion of NTM isolates obtained from all subjects who provided samples for mycobacterial culture increased over time. The proportion increased from 3.5% in 2000 to 6.3% in 2013, which was lower than that reported in our study (23). However, the study reported an increasing trend in prevalence (23). The prevalence of NTM isolated from sputum samples of patients with cystic fibrosis from Canada (6.1%) (25) and Israel (22.6%) (26) was lower than that reported in our study. Other studies reported the prevalence of NTM at 7.4 and 17.4% (27, 28).
It was reported that the NTM prevalence varies significantly between countries (6). All the previous studies used the culture method to detect NTM, the suggestion of The American Thoracic Society of repeat culture of sputum to establish pulmonary disease was mostly ignored, and NTM is often disregarded as a contaminant (3). Also, the lack of a definitive technique to identify NTM could contribute to the lower prevalence rates reported in earlier studies (21, 23, 24). The use of the advanced BACTEC technique with duplicate samples gave us the confidence that the positive samples could not be discarded as a contaminant, and it could increase the identification rate of NTM in our study.
Among different species, the M. avium complex (24.8%) was the predominant species identified, followed by M. kansasii (24%) and M. abscessus complex (20.2%) in our study. A recent study from South China, which described the epidemiology of pulmonary disease due to NTM, reported that M. avium complex (44.5%) was the predominant species identified among NTM, which is similar to our findings, but the identification rate was higher than in our study. The M. abscessus complex (40.5%) was the second most prevalent species identified, followed by M. kansasii (10.0%) and M. fortuitum (2.8%) (4). While in our study, M. kansasii (24%) was the second most species identified, followed by M. abscessus complex (20.2%). Nasiri et al. (24) from Iran reported M. simiae (38.7%) as the most predominant species identified, followed by M. fortuitum (19.3%) (24). In our study, M. simiae (16.3%) was the fourth most species identified, and M. fortuitum (8.5%) was the fifth most species identified. Jesudason et al. (21) from India reported that M. chelonae (46%) was the predominant NTM species isolated, and M. fortuitum (41%) was the second most NTM species isolated from various clinical samples (21).
Goswami et al. (22) from India reported M. fortuitum (4.61%) as the predominant species identified, although not predominant, while our study reported a higher rate (8.5%) of M. fortuitum. Cowman et al. (23) from the United Kingdom reported that M. abscessus was the predominant species identified among the younger age group, followed by M. avium complex and M. chelonae, while M. avium complex was the most predominant species identified in older age groups, followed by M. fortuitum, and M. xenopi (23). In our study, M. abscessus was the third common species identified; however, we could not identify any association with the age of patients. This is while M. avium complex was the most common species (32, 24.8%) identified, and of the 32 patients who were positive for M. avium complex, 25 patients were older than 50 years of age. The prevalence and predominance of different NTM species vary across different regions and studies. There was no similarity between the NTM species identified in our study and earlier reports (21, 23, 24). This could be due to the variation in the techniques used in different studies and the population included in the studies. These results suggest that there exists a clear geographical variation in the presence of different species of NTM. Thus, the species-level identification of NTM in every clinical setting is required for effective patient management.
In our study, 62.8% of our isolates were resistant to rifampicin. Rifampicin is the most common drug used for the treatment of tuberculosis. Rifampicin resistance was followed by resistance to levofloxacin (60.5%), ofloxacin (58.1%), and ethambutol (55.8%). Candido et al. (29) reported that amikacin and other aminoglycosides are commonly used for the treatment of bacterial infections, and M. abscessus isolates might show resistance to these drugs (29). In our study, 65.4% of the M. abscessus complex isolates were resistant to streptomycin, and 42.3% were resistant to amikacin. In our study, 56.7% of the isolates were MDR. Candido et al. (29) reported that 97.2% of their NTM isolates were resistant to five or more antibiotics, which is much higher than that reported in our study (29). A study from Iran, which isolated NTM from respiratory samples, reported that most of their NTM strains were resistant to multiple antibiotics; however, the study did not clearly mention the rate of MDR (30). As drug susceptibility is not routinely done in clinical settings, reports on MDR NTM are very scarce; hence, we could not extensively compare our MDR data with other reports.
We showed that 100% of the M. xenopi (n = 4) isolates were susceptible to streptomycin and amikacin; M. chelonae isolates (n = 2) were susceptible to ethambutol, streptomycin, and amikacin; M. fortuitum isolates (n = 11) were susceptible to amikacin, and M. gordonae isolates (n = 2) were susceptible to streptomycin. Another study from India reported that the majority (95.2%) of the M. fortuitum isolates were susceptible to amikacin, which is similar to our findings (22). In our study, 100% of the M. fortuitum and M. chelonae isolates were susceptible to amikacin, which is in complete agreement with that reported in India and Taiwan (22, 31). Similar to our findings, two other studies reported that 100% of the M. fortuitum isolates were susceptible to amikacin (32, 33). Cowman et al. (23) reported that except for streptomycin, most of the M. avium complex isolates were susceptible to isoniazid, rifampicin, and ciprofloxacin. The study also reported that M. kansasii showed high rates of susceptibility to all the tested antibiotics, and rifampicin resistance was rare (23).
In contrast, except for streptomycin (37.5%), more than 50% of our M. avium complex isolates were resistant to all other tested antibiotics. The resistance rate of M. kansasii in our study for different antibiotics tested ranged from 32.2 to 77.4 and 71.4%. Besides, 62.8% of our isolates were resistant to rifampicin. Goswami et al. (22) reported that all of their NTM isolates showed complete resistance to all tested antibiotics, except for streptomycin, and similar to our study, the majority of the isolates were resistant to all tested antibiotics (22). Antibiotic resistance to NTM greatly varies based on species and geographical location (22).
The use of anti-tuberculosis drugs in specific clinical settings plays a major role in determining antibiotic resistance. A study from China reported high drug resistance by NTM to first-line anti-TB drugs; 30.7% of the isolates were suspected to be of MDR-TB cases and 4.0% of TB re-treatment cases; the study suggests that pulmonary NTM infections could cause substantial difficulties in the clinical management of NTM and MDR-TB diseases in China (13). A predominant number of our isolates were resistant to rifampicin and routinely used anti-tuberculosis drugs, indicating that NTM has been exposed much to the drug due to overuse, which led to selective pressure and might have contributed to the commonly used anti-tuberculosis drugs.
The limitations of the study include the non-availability of clinical and radiological data, which could have revealed the clinical significance of the isolates; with the limited data available from the microbiology, we could not determine additional evidence of NTM infections such as positive cultures from other centers and the history of the disease condition.
5.1. Conclusions
Mycobacterium avium complex was the predominant species identified, and the majority of the organisms were resistant to commonly used anti-tuberculosis drugs. The high prevalence of NTM and drug resistance towards the tested antibiotics suggests that NTM can no more be ignored as a contaminant, reiterating the need for periodic surveillance and species-specific treatments for effective management of diseases caused by NTM.
References
-
1.
Rindi L, Garzelli C. Increase in non-tuberculous mycobacteria isolated from humans in Tuscany, Italy, from 2004 to 2014. BMC Infect Dis. 2016;16:44. [PubMed ID: 26831721]. [PubMed Central ID: PMC4736237]. https://doi.org/10.1186/s12879-016-1380-y.
-
2.
Ratnatunga CN, Lutzky VP, Kupz A, Doolan DL, Reid DW, Field M, et al. The rise of non-tuberculosis mycobacterial lung disease. Front Immunol. 2020;11:303. [PubMed ID: 32194556]. [PubMed Central ID: PMC7062685]. https://doi.org/10.3389/fimmu.2020.00303.
-
3.
Griffith DE, Aksamit T, Brown-Elliott BA, Catanzaro A, Daley C, Gordin F, et al. An official ATS/IDSA statement: Diagnosis, treatment, and prevention of nontuberculous Mycobacterial diseases. Am J Respir Crit Care Med. 2007;175(4):367-416. [PubMed ID: 17277290]. https://doi.org/10.1164/rccm.200604-571ST.
-
4.
Tan Y, Su B, Shu W, Cai X, Kuang S, Kuang H, et al. Epidemiology of pulmonary disease due to nontuberculous mycobacteria in Southern China, 2013-2016. BMC Pulm Med. 2018;18(1):168. [PubMed ID: 30413193]. [PubMed Central ID: PMC6230232]. https://doi.org/10.1186/s12890-018-0728-z.
-
5.
Kim SY, Han SA, Kim DH, Koh WJ. Nontuberculous mycobacterial lung disease: Ecology, microbiology, pathogenesis, and antibiotic resistance mechanisms. Precis Future Med. 2017;1(3):99-114. https://doi.org/10.23838/pfm.2017.00135.
-
6.
Adjemian J, Olivier KN, Seitz AE, Holland SM, Prevots DR. Prevalence of nontuberculous mycobacterial lung disease in U.S. Medicare beneficiaries. Am J Respir Crit Care Med. 2012;185(8):881-6. [PubMed ID: 22312016]. [PubMed Central ID: PMC3360574]. https://doi.org/10.1164/rccm.201111-2016OC.
-
7.
Prevots DR, Shaw PA, Strickland D, Jackson LA, Raebel MA, Blosky MA, et al. Nontuberculous mycobacterial lung disease prevalence at four integrated health care delivery systems. Am J Respir Crit Care Med. 2010;182(7):970-6. [PubMed ID: 20538958]. [PubMed Central ID: PMC2970866]. https://doi.org/10.1164/rccm.201002-0310OC.
-
8.
Donohue MJ, Wymer L. Increasing prevalence rate of nontuberculous mycobacteria infections in five states, 2008-2013. Ann Am Thorac Soc. 2016;13(12):2143-50. [PubMed ID: 27681202]. https://doi.org/10.1513/AnnalsATS.201605-353OC.
-
9.
Shah NM, Davidson JA, Anderson LF, Lalor MK, Kim J, Thomas HL, et al. Pulmonary Mycobacterium avium-intracellulare is the main driver of the rise in non-tuberculous mycobacteria incidence in England, Wales and Northern Ireland, 2007-2012. BMC Infect Dis. 2016;16:195. [PubMed ID: 27154015]. [PubMed Central ID: PMC4858927]. https://doi.org/10.1186/s12879-016-1521-3.
-
10.
Wu J, Zhang Y, Li J, Lin S, Wang L, Jiang Y, et al. Increase in nontuberculous mycobacteria isolated in Shanghai, China: Results from a population-based study. PLoS One. 2014;9(10). e109736. [PubMed ID: 25330201]. [PubMed Central ID: PMC4199589]. https://doi.org/10.1371/journal.pone.0109736.
-
11.
Abubakar I, Gupta RK, Rangaka MX, Lipman M. Update in tuberculosis and nontuberculous mycobacteria 2017. Am J Respir Crit Care Med. 2018;197(10):1248-53. [PubMed ID: 29537298]. https://doi.org/10.1164/rccm.201801-0106UP.
-
12.
Johnson MM, Odell JA. Nontuberculous mycobacterial pulmonary infections. J Thorac Dis. 2014;6(3):210-20. [PubMed ID: 24624285]. [PubMed Central ID: PMC3949190]. https://doi.org/10.3978/j.issn.2072-1439.2013.12.24.
-
13.
Jing H, Wang H, Wang Y, Deng Y, Li X, Liu Z, et al. Prevalence of nontuberculous mycobacteria infection, China, 2004-2009. Emerg Infect Dis. 2012;18(3):527-8. [PubMed ID: 22376989]. [PubMed Central ID: PMC3309567]. https://doi.org/10.3201/eid1803.110175.
-
14.
Caviedes L, Lee TS, Gilman RH, Sheen P, Spellman E, Lee EH, et al. Rapid, efficient detection and drug susceptibility testing of Mycobacterium tuberculosis in sputum by microscopic observation of broth cultures. The tuberculosis working group in Peru. J Clin Microbiol. 2000;38(3):1203-8. [PubMed ID: 10699023]. [PubMed Central ID: PMC86377]. https://doi.org/10.1128/JCM.38.3.1203-1208.2000.
-
15.
World Health Organization. Noncommercial culture and drug-susceptibility testing methods for screening patients at risk for multidrug-resistant tuberculosis: Policy statement. Geneva, Switzerland: World Health Organization; 2011.
-
16.
Wang L, Mohammad SH, Chaiyasirinroje B, Li Q, Rienthong S, Rienthong D, et al. Evaluating the Auto-MODS assay, a novel tool for tuberculosis diagnosis for use in resource-limited settings. J Clin Microbiol. 2015;53(1):172-8. [PubMed ID: 25378569]. [PubMed Central ID: PMC4290917]. https://doi.org/10.1128/JCM.01946-14.
-
17.
Kam KM. Microscopic observation drug susceptibility (MODS): Where are we going? Int J Tuberc Lung Dis. 2014;18(2):127. [PubMed ID: 24429301]. https://doi.org/10.5588/ijtld.13.0777.
-
18.
Tripathi K, Tripathi PC, Nema S, Shrivastava AK, Dwiwedi K, Dhanvijay AK. Modified Petroff’s method: An excellent simplified decontamination technique in comparison with Petroff’s method. Int J Recent Trends Sci Tech. 2014;10(3):461-4.
-
19.
Moore DA, Evans CA, Gilman RH, Caviedes L, Coronel J, Vivar A, et al. Microscopic-observation drug-susceptibility assay for the diagnosis of TB. N Engl J Med. 2006;355(15):1539-50. [PubMed ID: 17035648]. [PubMed Central ID: PMC1780278]. https://doi.org/10.1056/NEJMoa055524.
-
20.
Stinson KW, Eisenach K, Kayes S, Matsumoto M, Siddiqi S, Nakashima S. Mycobacteriology Laboratory Manual, global laboratory initiative advancing TB diagnosis. 1st ed. Geneva, Switzerland: Global Laboratory Initiative Advancing Tb Diagnosis; 2014.
-
21.
Jesudason MV, Gladstone P. Non tuberculous mycobacteria isolated from clinical specimens at a tertiary care hospital in South India. Indian J Med Microbiol. 2005;23(3):172-5. [PubMed ID: 16100423]. https://doi.org/10.4103/0255-0857.16589.
-
22.
Goswami B, Narang P, Mishra PS, Narang R, Narang U, Mendiratta DK. Drug susceptibility of rapid and slow growing non-tuberculous mycobacteria isolated from symptomatics for pulmonary tuberculosis, Central India. Indian J Med Microbiol. 2016;34(4):442-7. [PubMed ID: 27934821]. https://doi.org/10.4103/0255-0857.195375.
-
23.
Cowman S, Burns K, Benson S, Wilson R, Loebinger MR. The antimicrobial susceptibility of non-tuberculous mycobacteria. J Infect. 2016;72(3):324-31. [PubMed ID: 26723913]. https://doi.org/10.1016/j.jinf.2015.12.007.
-
24.
Nasiri MJ, Dabiri H, Fooladi AAI, Amini S, Hamzehloo G, Feizabadi MM. High rates of nontuberculous mycobacteria isolation from patients with presumptive tuberculosis in Iran. New Microbes New Infect. 2018;21:12-7. [PubMed ID: 29188063]. [PubMed Central ID: PMC5695646]. https://doi.org/10.1016/j.nmni.2017.08.008.
-
25.
Radhakrishnan DK, Yau Y, Corey M, Richardson S, Chedore P, Jamieson F, et al. Non-tuberculous mycobacteria in children with cystic fibrosis: Isolation, prevalence, and predictors. Pediatr Pulmonol. 2009;44(11):1100-6. [PubMed ID: 19830845]. https://doi.org/10.1002/ppul.21106.
-
26.
Levy I, Grisaru-Soen G, Lerner-Geva L, Kerem E, Blau H, Bentur L, et al. Multicenter cross-sectional study of nontuberculous mycobacterial infections among cystic fibrosis patients, Israel. Emerg Infect Dis. 2008;14(3):378-84. [PubMed ID: 18325250]. [PubMed Central ID: PMC2570835]. https://doi.org/10.3201/eid1403.061405.
-
27.
Chakrabarti A, Sharma M, Dubey ML. Isolation rates of different mycobacterial species from Chandigarh (North India). Indian J Med Res. 1990;91:111-4. [PubMed ID: 2111799].
-
28.
Karak K, Bhattacharyya S, Majumdar S, De PK. Pulmonary infection caused by mycobacteria other than M. tuberculosis in and around Calcutta. Indian J Pathol Microbiol. 1996;39(2):131-4. [PubMed ID: 9401242].
-
29.
Candido PH, Nunes Lde S, Marques EA, Folescu TW, Coelho FS, de Moura VC, et al. Multidrug-resistant nontuberculous mycobacteria isolated from cystic fibrosis patients. J Clin Microbiol. 2014;52(8):2990-7. [PubMed ID: 24920766]. [PubMed Central ID: PMC4136125]. https://doi.org/10.1128/JCM.00549-14.
-
30.
Mortazavi Z, Bahrmand A, Sakhaee F, Doust RH, Vaziri F, Siadat SD, et al. Evaluating the clinical significance of nontuberculous mycobacteria isolated from respiratory samples in Iran: An often overlooked disease. Infect Drug Resist. 2019;12:1917-27. [PubMed ID: 31308711]. [PubMed Central ID: PMC6613451]. https://doi.org/10.2147/IDR.S214181.
-
31.
Yang SC, Hsueh PR, Lai HC, Teng LJ, Huang LM, Chen JM, et al. High prevalence of antimicrobial resistance in rapidly growing mycobacteria in Taiwan. Antimicrob Agents Chemother. 2003;47(6):1958-62. [PubMed ID: 12760874]. [PubMed Central ID: PMC155839]. https://doi.org/10.1128/aac.47.6.1958-1962.2003.
-
32.
Gayathri R, Therese KL, Deepa P, Mangai S, Madhavan HN. Antibiotic susceptibility pattern of rapidly growing mycobacteria. J Postgrad Med. 2010;56(2):76-8. [PubMed ID: 20622384]. https://doi.org/10.4103/0022-3859.65278.
-
33.
Swenson JM, Thornsberry C, Silcox VA. Rapidly growing mycobacteria: Testing of susceptibility to 34 antimicrobial agents by broth microdilution. Antimicrob Agents Chemother. 1982;22(2):186-92. [PubMed ID: 6927280]. [PubMed Central ID: PMC183707]. https://doi.org/10.1128/aac.22.2.186.