Abstract
Background:
Carbapenem-resistant Klebsiella pneumoniae (CR-KP), known as a significant public health threat, is the most common causative agent of nosocomial and community-acquired infections.Objectives:
This study aimed to evaluate resistance to carbapenems and determine the prevalence of carbapenemase genes and multilocus sequence typing (MLST) of K. pneumoniae clinical isolates.Methods:
One-hundred K. pneumoniae isolates were evaluated. The minimum inhibitory concentrations (MIC) of imipenem and meropenem were assessed by the broth microdilution method. Multiplex-polymerase chain reaction (PCR) was applied to detect 11 carbapenemase-encoding genes belonging to different classes. The alleles and sequence types (ST) of three isolates were identified by MLST.Results:
The MIC of carbapenems for the isolates ranged from 0.062 to 32 µg/mL. Overall, resistance rates to imipenem and meropenem were reported 11% and 34%, respectively. The bla IMP gene was the most abundant (78.4%), followed by bla OXA-48 (48.6%), bla GIM (27%), bla KPC (27%), bla SIM (21.6%), bla BIC (21.6%), bla NDM (16.2%), bla AIM (16.2%), bla VIM (16.2%), bla DIM (8.1%), and bla SPM (8.1%). The co-existence of carbapenemase genes was observed in 81.8% of the isolates. A positive relationship was found between the presence of bla NDM and bla SIM and resistance to imipenem. Multilocus sequence typing results showed three different sequence types, including ST14, ST5188, and ST1861.Conclusions:
This study revealed a high prevalence of CR-KP isolates that suggests a high risk of horizontal gene transfer and potential to spread resistance among other strains. Since STs are reported for the first time in Iran, they can be considered as emerging strains.Keywords
Klebsiella pneumoniae Multilocus Sequence Typing Carbapenemase Imipenem Meropenem
1. Background
Klebsiella pneumoniae is an opportunistic pathogen and one of the most important bacteria in hospital-acquired infections, causing soft tissue infections, septicemia, pneumonia, and urinary tract infections in patients admitted to hospital wards (1). Beta-lactam antibiotics are primarily used for the treatment of Gram-negative infections (1). Of the beta-lactam antibiotics, carbapenems have the broadest spectrum of inhibitory activity against Gram-positive and Gram-negative bacteria. (1). Although resistance to carbapenems is still rare, it is emerging and poses a significant threat to the management of multidrug-resistant isolates in many clinics and hospitals. Currently, several carbapenems are used in clinical practice, including imipenem, meropenem, ertapenem, doripenem, panipenem, biopenem, reopenem, and faropenem (2, 3). However, in the last decade, the emergence and spread of multidrug-resistant strains among isolates have become a major therapeutic problem, requiring microbiological and epidemiological interventions (4).
Carbapenem resistance in K. pneumoniae isolates is mediated by various mechanisms such as increased production of efflux pumps, reduced outer membrane permeability, changing tendency of the proteins bound to carbapenems, decreased production of porins, etc. However, in K. pneumoniae isolates, the major mechanism of acquired beta-lactam resistance is the production of beta-lactamases (3). Beta-lactamases are classified based on both structural (Bosch method) and functional (Ambler method) properties. The simplest classification of beta-lactamases is by protein sequence, whereby they are classified into the four molecular classes of A, B, C, and D. Clinically-relevant carbapenemases belong either to Ambler class A, B, and D or to Bosch groups 1, 2f, 2df, 3a, and 3b. The Ambler class A and Group 2f include members of the IMI, SME, GES, KPC, and NMC enzymes.
The Ambler class B and Group 3a and 3b include IMP, VIM, GIM, SPM, CcrA, IND-1, CphA, and Sfh-1 enzymes. Finally, OXA enzymes were placed in class D and group 2df (5). The K. pneumoniae carbapenemase (KPC) enzyme is the most concerning Ambler class A because of its location on self-conjugative plasmids and ability to transfer resistance among Enterobacteriaceae. This enzyme is able to hydrolyze penicillins, classical cephalosporins, monobactam, and all carbapenems such as imipenem, meropenem, ertapenem, and doripenem, and it is weakly inhibited by clavulanic acid and tazobactam (6). The enzymes of the VIM, IMP, and NDM groups are in Ambler class B, and their genes are located on the plasmid. These enzymes cannot be physically bound to the beta-lactam substrate and therefore escape the action of beta-lactamase inhibitors such as clavulanic acid and sulbactam and are able to bind to all beta-lactam classes, except monobactam (7, 8). OXA-type carbapenemase enzymes are also in the D class of Ambler and are known as oxacillinases due to their accelerated hydrolysis of classical penicillins. These enzymes are divided into nine groups based on amino acid similarity, and their genes are located on chromosomes and plasmids. The bla OXA-48 enzyme, which is in the sixth group of OXA enzymes, is one of the most important and common enzymes in the development of carbapenem resistance, and its gene is on the plasmid (9).
The ability to accurately characterize the pathogenic strains is crucial for epidemiological surveillance and the development of public health control strategies (10). To understand the epidemiological characterization and prevent the spread of resistant strains of bacteria, researchers classify them by molecular techniques such as multilocus sequence typing (MLST). In the MLST technique, isolates of bacterial species are characterized using the partial sequence analysis of housekeeping genes. For each housekeeping gene, different sequences in a bacterial species are specified as distinct alleles, and the alleles at each of the seven loci determine the sequence type (ST) of each isolate (11). Multilocus sequence typing possesses a number of advantages over other typing methods: 1) it utilizes sequence data and thus can detect changes at DNA level, which are not distinguished by other phenotypic methods such as serotyping and by multilocus enzyme electrophoresis (MLEE); 2) Multilocus sequence typing does not require live bacterial isolates and can be directly performed on clinical samples of patients undergoing antimicrobial therapy, from which live bacteria may be difficult to isolate; and 3) The data obtained by MLST are fully portable between laboratories and can be easily shared all around the world through the Internet (12).
2. Objectives
This study aimed to evaluate resistance to carbapenems and determine the prevalence of carbapenemase genes and MLST of K. pneumoniae clinical isolates.
3. Methods
3.1. Collection and Identification of Isolates
The present study was a descriptive cross-sectional study performed on 100 clinical isolates of K. pneumoniae. These bacteria were collected from urine samples of patients admitted to Milad Hospital in Tehran from October 2018 to August 2019 and were confirmed as K. pneumoniae using standard microbiological tests (13).
3.2. Minimum Inhibitory Concentration and Minimum Bactericidal Concentration Determinations
All strains were cultured, and antibiotic sensitivity tests were performed by microdilution broth methods using the Clinical and Laboratory Standards Institute (CLSI) guidelines (14). The antimicrobial agents included in this study were imipenem and meropenem (Sigma-Aldrich). Dilutions of carbapenems (0.125 - 256 μg/mL) in Mueller Hinton Broth (MHB) (Difco, Sparks, Md) and dilutions of the bacterial suspension (OD = 0.8 to 1 at 625 nm) were prepared. Briefly, 100 μL of the bacterial suspension and 100 μL of imipenem or meropenem at different concentrations were poured into each well of microplates. Wells containing 100 μL of each antibiotic (256 μg/mL) and 100 μL of MHB were considered as a negative control.
For positive control, 100 μL of bacterial suspension was added to 100-μL MHB. The microplates were incubated at 37°C for 18 - 24 h. The minimum inhibitory concentration (MIC) was considered as the lowest amount of the antibacterial agents that inhibited the visible growth of bacteria, and the results were interpreted according to the MIC Breakpoints for Enterobacteriaceae. The breakpoints assigned by CLSI for imipenem and meropenem MICs against Enterobacteriaceae are ≤ 1 μg/ mL (susceptible), 2 μg/ mL (intermediate), and ≥ 4 μg/ mL (resistant). Afterward, 5 μl of the last three wells without visible growth were cultured on tryptic soy agar (Merck, Germany) plates and incubated at 37°C for 24 h. The minimum bactericidal concentration (MBC) was defined as the lowest concentration at which no bacterial growth was observed.
3.3. Molecular Detection of Carbapenemase Genes
First, DNA extraction was performed from overnight cultures of the isolates in tryptic soy broth (TSB) (Merck, Germany) by the boiling method as described previously (15). For the identification of carbapenemase genes, multiplex polymerase chain reaction (PCR) was run for each isolate to evaluate the presence of the following genetic targets: class A beta-lactamase genes (bla KPC and bla BIC), class B beta-lactamase genes (bla SPM, bla AIM, bla IMP, bla VIM, bla NDM , bla SIM, bla GIM, and bla DIM), and class D beta-lactamase genes (bla OXA) (Table 1). The reaction mixture (25 µL) contained 12.5 µL of the 2X Master Mix RED (Amplicon), 0.5 µL (25 pmol) of each primer, 2.5 μL (300 ng) of extracted DNA, and 9 μL of deionized water (SinaClon, Iran). Thermal cycler conditions were as follows: primary denaturation at 94°C for 10 min, followed by denaturation at 94°C for 30 s, annealing at 52°C for 40 s, extension at 72°C for 50 s (35 cycles), and a final extension at 72°C for 5 min. Finally, PCR products were analyzed by electrophoresis on 1% agarose gel in TBE buffer containing DNA Safe Stain (5 µL/L).
| Gene | Beta-Lactamases | Primer Sequence | Size (bp) |
|---|---|---|---|
| bla KPC | Class A | 798 | |
| F | CGTCTAGTTCTGCTGTCTTG | ||
| R | CTTGTCATCCTTGTTAGGCG | ||
| bla BIC | 537 | ||
| F | TATGCAGCTCCTTTAAGGGC | ||
| R | TCATTGGCGGTGCCGTACAC | ||
| bla IMP | Class B | 232 | |
| F | GGAATAGAGTGGCTTAAYTCTC | ||
| R | GGTTTAAYAAAACAACCACC | ||
| blaVIM | 390 | ||
| F | GATGGTGTTTGGTCGCATA | ||
| R | CGAATGCGCAGCACCAG | ||
| blaNDM | 621 | ||
| F | GGTTTGGCGATCTGGTTTTC | ||
| R | CGGAATGGCTCATCACGATC | ||
| blaSPM | 271 | ||
| F | AAAATCTGGGTACGCAAACG | ||
| R | ACATTATCCGCTGGAACAGG | ||
| blaAIM | 322 | ||
| F | CTGAAGGTGTACGGAAACAC | ||
| R | GTTCGGCCACCTCGAATTG | ||
| blaGIM | 477 | ||
| F | TCGACACACCTTGGTCTGAA | ||
| R | AACTTCCAACTTTGCCATGC | ||
| blaSIM | 570 | ||
| F | TACAAGGGATTCGGCATCG | ||
| R | TAATGGCCTGTTCCCATGTG | ||
| bla DIM | 699 | ||
| F | GCTTGTCTTCGCTTGCTAACG | ||
| R | CGTTCGGCTGGATTGATTTG | ||
| bla OXA-48 | Class D | 438 | |
| F | GCGTGGTTAAGGATGAACAC | ||
| R | CATCAAGTTCAACCCAACCG |
3.4. Multilocus Sequence Typing
Multilocus sequence typing was carried out as described previously (17). The internal fragments of seven housekeeping genes (i.e., rpoB, gapA, mdh, pgi, phoE, infB, and tonB) were amplified and sequenced using specific primers by referring to online MLST database (Table 2). The polymerase chain reaction was carried out in a 50-μL reaction mixture containing 25 µL of the 2x Master Mix RED (Amplicon), 1 μL (25 pmol) of each of the forward and reverse primers, 5 μL (600 ng) of extracted DNA, and 18 μL of nuclease free water (SinaClon, Iran). The PCR program was as follows: initial denaturation at 94°C for 2 min followed by 35 cycles of denaturation at 94°C for 20 s, annealing at 45 - 60°C for 30 s (based on each gene), extension at 72°C for 30 s, and a final extension at 72°C for 5 min. The PCR products were sequenced by CinnaGen Company in South Korea. The nucleotide sequences of the housekeeping genes were submitted to the K. pneumoniae MLST Database (https://bigsdb.pasteur.fr/klebsiella/klebsiella.html) to detect the allele numbers and STs. Also, the clonal complex (CC) of all the three isolates was determined using the goeBURST algorithm, version 1.2.1.
| Locus | Primer sequence | Size (bp) | No. of Alleles |
|---|---|---|---|
| rpoB | 501 | 8 | |
| F | GGCGAAATGGCWGAGAACCA | ||
| R | GAGTCTTCGAAGTTGTAACC | ||
| gapA | 450 | 6 | |
| F | TGAAATATGACTCCACTCACGG | ||
| R | CTTCAGAAGCGGCTTTGATGGCTT | ||
| mdh | 477 | 10 | |
| F | CCCAACTCGCTTCAGGTTCAG | ||
| R | CCGTTTTTCCCCAGCAGCAG | ||
| pgi | 432 | 6 | |
| F | GAGAAAAACCTGCCTGTACTGCTGGC | ||
| R | CGCGCCACGCTTTATAGCGGTTAAT | ||
| phoE | 420 | 14 | |
| F | ACCTACCGCAACACCGACTTCTTCGG | ||
| R | TGATCAGAACTGGTAGGTGAT | ||
| infB | 318 | 10 | |
| F | CTCGCTGCTGGACTATATTCG | ||
| R | CGCTTTCAGCTCAAGAACTTC | ||
| tonB | 414 | 21 | |
| F | CTTTATACCTCGGTACATCAG GTT | ||
| R | ATTCGCCGGCTGRGCRGAGAG |
3.5. Statistical Analysis
The association of each carbapenem-resistant gene with resistance to imipenem or meropenem was specified by Pearson’s correlation coefficient, chi-square test, and Fisher’s exact test using SPSS, version 20. A P-value of less than 0.05 was considered significant.
4. Results
4.1. Determination of Minimum Inhibitory Concentration and Minimum Bactericidal Concentration
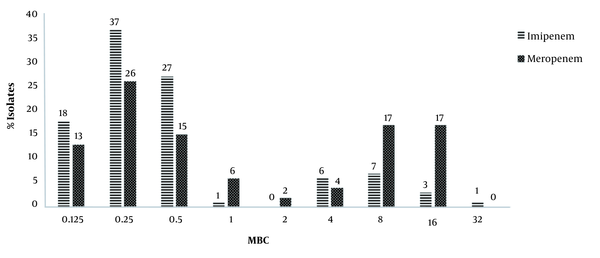
Evaluation of drug susceptibility by the broth microdilution method showed that the MICs of imipenem and meropenem for respectively 83 (83%) and 60 (60 %) isolates were less than 1 μg/mL, which were considered as sensitive strains. Also, 11 (11%) and 34 (34%) isolates were resistant to imipenem and meropenem, respectively, and 6 (6%) isolates showed intermediate susceptibility to both antibiotics. Also, the amount of MBC of imipenem and meropenem ranged from 0.125 to 32 µg/mL (Figure 1).
Minimum bactericidal concentration determination results of carbapenems by the broth microdilution method
4.2. Carbapenemase Resistance Genes
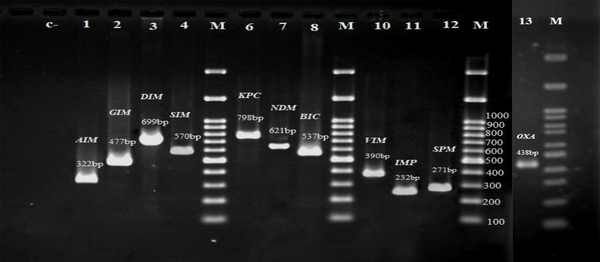
A total of 37 K. pneumoniae were resistant to carbapenems (meropenem and/ or imipenem) and these isolates were tested for the presence of carbapenem resistance genes. In the electrophoresis of the PCR products, the expected bands for each gene were observed and recorded, which are shown in Figure 2. Of the 11 carbapenemase genes identified in the 37 isolates of K. pneumoniae, the highest frequency was related to the bla IMP gene (78.4%), followed by bla OXA-48 (48.6%), bla GIM (27%), bla KPC (27%), bla SIM (21.6%), bla BIC (21.6%), bla NDM (16.2%), bla AIM (16.2%), bla VIM (16.2%), bla DIM (8.1%), and bla SPM (8.1%). Gene frequency analysis revealed that all the 37 isolates carried at least one of the studied carbapenemase genes. In general, seven isolates carried at least one gene, ten isolates two genes, eight isolates three genes, six isolates four genes, five isolates five genes, and one isolate carried seven genes simultaneously.
Electrophoresis of the polymerase chain reaction products of carbapenemase genes. C- Negative control, M: 100 bp DNA ladder, Lane 1-13: PCR product of carbapenemase genes
4.3. Multilocus Sequence Typing
In the present study, STs of three carbapenem-resistant isolates were identified by the MLST method. These isolates showed simultaneous resistance against imipenem and meropenem and had the highest MIC against the antibiotics tested (Table 3). After the sequencing of the seven housekeeping genes, the alleles and STs were assigned according to the MLST international scheme of the Institut Pasteur, Paris, France. The rpoB allele was similar in all three isolates, and the infB allele was identical in both isolates 4 and 11. eBURST analysis showed that all three isolates were in different clonal complexes (Table 3).
MIC, Resistance Genes, Allele Numbers, and ST Type of Three Isolates of Klebsiella pneumoniae
| Isolates | MIC(μL/mL) | Carbapenemase Genes | Allele Numbers | ST | CC | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Imi | Mer | gapA | infB | mdh | pgi | phoE | rpoB | tonB | ||||
| K4 | 4 | 8 | bla IMP, bla KPC, bla OXA-48 | 276 | 1 | 11 | 20 | 3 | 1 | 13 | 5188 | 8 |
| K11 | 8 | 4 | bla IMP, bla AIM, bla SIM, bla NDM, bla OXA-48 | 2 | 1 | 166 | 46 | 9 | 1 | 12 | 1861 | 3 |
| K13 | 16 | 8 | bla IMP, bla SIM, bla KPC,bla OXA-48 | 1 | 6 | 1 | 1 | 1 | 1 | 1 | 14 | 41 |
5. Discussion
Beta-lactams, including carbapenems, are widely used in the treatment of Gram-negative bacterial infections. In recent years, carbapenems, including imipenem and meropenem, have become the most effective drugs for the treatment of serious infections in hospitalized patients (18). However, resistance to these drugs has been reported by researchers in many cases, which has led to further attention to this issue. Carbapenem-resistant K. pneumoniae, due to the narrow therapeutic options and the high mortality rate, poses significant public health challenges worldwide (19). In the present study, the evaluation of drug resistance to carbapenems by the broth microdilution method showed that resistance rates to imipenem and meropenem were 11% and 34%, respectively. In studies conducted in other regions of Iran, resistance to carbapenems has been usually lower than our findings. In this case, we can refer to the studies of Rahbar et al. in 2008 (20), Mansouri and Abbasi in 2010 (21), and Sultan Dalal et al. in 2012 (22), who noted that resistance to imipenem and meropenem was less than 2%.
In studies by Amin et al. in Pakistan (23), Ishii et al. in Japan (24), and Al-Shara and Mohammad Abdullah in Jordan (25), imipenem as an effective drug has been introduced in the treatment of Klebsiella infection. However, in recent years, resistance to carbapenems has increased. Gurung et al. in Nepal reported that resistance to imipenem and meropenem among clinical isolates of K. pneumoniae was 16% (26). Soroush and Ghane (2017) reported that 25% of K. pneumoniae isolates from ICU patients were resistant to imipenem (27). In another study conducted by Khairy et al. (2020), it was shown that out of 42 isolates, 57% of the isolates were resistant to meropenem and 13% were resistant to imipenem (28). In the present study, resistance to carbapenems was moderate, but it could be a concern due to the placement of resistance genes on the mobilizable plasmids and their easy transfer between isolates.
Carbapenem resistance in K. pneumoniae is provided by several mechanisms, but many studies have suggested the presence of carbapenemase enzymes as one of the main reason for resistance to these drugs (29). In the present study, 11 carbapenemase genes from the three classes of A, B, and D beta-lactamase were detected by the PCR method, and the bla IMP gene was found to be more prevalent (87%). A review of other studies showed that the bla IMP gene had a higher frequency (30-32), which was in line with the results of this study. It should be noted that all carbapenem-non-susceptible K. penomoniae carried at least one of the carbapenemase genes. Furthermore, the isolates showing resistance to both carbapenem harbored four or more carbapenem resistance genes simultaneously. These findings indicate that carbapenemase enzymes play an important role in resistance to carbapenems. However, studies have indicated that other mechanisms might alternatively contribute to carbapenem resistance, including deficiency in porin, decreased expression of outer membrane proteins, or production of oxacillinase (33). Carbapenemase genes have been reported in other Enterobacteriaceae and Gram-negative bacteria, and their placement on the plasmid increases the risk of their spread.
bla KPC gene, one of the beta-lactamase group A genes, has been reported as the most important gene involved in the development of carbapenem resistance. In a study in Brazil, Pavez et al. attributed the resistance of a K. pneumoniae isolate to imipenem to the enzyme KPC (34). Bratu in 2007 and Nordmann in 2011 in the United States reported the KPC enzyme as the cause of resistance of an Escherichia coli isolate to carbapenems (35, 36). In the present study, the bla KPC gene had a frequency of 27% among the K. pneumoniae isolates, which was moderate compared to other studies conducted in other countries (31). In China, most CR-KP isolates developed carbapenem resistance by producing KPCs 9-11 (37). Klebsiella pneumoniae carbapenemase-producing bacteria are clinically significant due to the narrow therapeutic options available and the high mortality rate caused by these bacterial infection (37).
Previous studies proved the inactivation of carbapenems such as imipenem, meropenem doripenem, and ertapenem by metallo-beta lactamase enzymes. It has been reported that bla NDM is the main mechanism of resistance of Gram-negative bacteria to carbapenems (38, 39). Similarly, in this study, we found a significant correlation between imipenem resistance and the carriage of both bla SIM and bla NDM genes (P = 0.001 and P = 0.016, respectively). However, no relationship was observed between resistance to carbapenems and other tested genes (P > 0.05). Therefore, it can be suggested that the bla SIM and bla NDM genes were among the main factors in the development of carbapenem resistance among our isolates. However, other mechanisms of resistance, such as deficiency or decreased expression of porins, must also be investigated.
In this study, to identify the epidemiological characteristics of strains, three isolates were classified by the MLST method. Multilocus sequence typing results showed that STs of isolates were different (ST5188, ST1861, ST14) (https://bigsdb.pasteur.fr/klebsiella/klebsiella.html). Klebsiella pneumoniae ST14, resistant to both carbapenem and colistin, was previously described in South Korea (40). ST 14 was also reported in hospitals from Dubai, United Arab Emirates, which produced OXA-48-type and NDM carbapenemases (41). Klebsiella pneumoniae ST14 obtained in our study was highly resistant to imipenem and meropenem. In addition to bla KPC and bla OXA-48, our isolate harbored bla IMP and bla SIM and belonged to the clonal complex 41 (CC 41). However, ST 5188 and ST 1861 belonged to the CC8 and CC3, respectively. These two STs have a low prevalence in the world. The latter was first described from a bacteremic liver abscess caused by a hypervirulent K. pneumoniae in Italy (42).
Although this strain was first described as a multi susceptible strain, ST 1861 reported in this study was resistant to both imipenem and meropenem and harbored bla IMP, bla AIM, bla SIM, bla NDM, and bla OXA-48 resistance genes. The occurrence of such isolates with a large number of carbapenemase genes could be dangerous to public health in the future, indicating an urgent need for epidemiologic surveillance and improved clinical awareness. Also, these types are reported for the first time in Iran and can be considered as emerging strains. In this case, Meng et al. (2019) reported that ST11 is the epidemic sequence type of KPC-producing K. pneumoniae in China that contributed to the spread of antibiotic resistance in hospitals (37).
5.1. Conclusions
Resistance of K. pneumoniae to carbapenems through the mechanism of carbapenemase production is becoming increasingly distressing. In the present study, out of 37 carbapenem-resistant isolates identified by phenotypic method, all the isolates carried at least one of the carbapenemase genes, and the bla NDM and bla SIM genes played an important role in resistance to carbapenems. The emergence of new ST types of K. pneumoniae with high drug resistance to carbapenems could also signal the spread of such strains in the future. Therefore, the rapid identification of metallo-beta-lactamase-producing K. pneumoniae isolates is necessary to prevent the further spread of infection by these organisms.
References
-
1.
Podschun R, Ullmann U. Klebsiella spp. as nosocomial pathogens: epidemiology, taxonomy, typing methods, and pathogenicity factors. Clin Microbiol Rev. 1998;11(4):589-603. [PubMed ID: 9767057]. [PubMed Central ID: PMC88898].
-
2.
Nicolau DP. Carbapenems: a potent class of antibiotics. Expert Opin Pharmacother. 2008;9(1):23-37. [PubMed ID: 18076336]. https://doi.org/10.1517/14656566.9.1.23.
-
3.
Codjoe FS, Donkor ES. Carbapenem resistance: A review. Med Sci (Basel). 2017;6(1). [PubMed ID: 29267233]. [PubMed Central ID: PMC5872158]. https://doi.org/10.3390/medsci6010001.
-
4.
Wroblewska MM, Towner KJ, Marchel H, Luczak M. Emergence and spread of carbapenem-resistant strains of Acinetobacter baumannii in a tertiary-care hospital in Poland. Clin Microbiol Infect. 2007;13(5):490-6. [PubMed ID: 17331123]. https://doi.org/10.1111/j.1469-0691.2007.01694.x.
-
5.
Queenan AM, Bush K. Carbapenemases: the versatile beta-lactamases. Clin Microbiol Rev. 2007;20(3):440-58. [PubMed ID: 17630334]. [PubMed Central ID: PMC1932750]. https://doi.org/10.1128/CMR.00001-07.
-
6.
Bush K. Past and present perspectives on beta-lactamases. Antimicrob Agents Chemother. 2018;62(10). [PubMed ID: 30061284]. [PubMed Central ID: PMC6153792]. https://doi.org/10.1128/AAC.01076-18.
-
7.
Bush K, Bradford PA. Interplay between beta-lactamases and new beta-lactamase inhibitors. Nat Rev Microbiol. 2019;17(5):295-306. [PubMed ID: 30837684]. https://doi.org/10.1038/s41579-019-0159-8.
-
8.
Yigit H, Queenan AM, Anderson GJ, Domenech-Sanchez A, Biddle JW, Steward CD, et al. Novel carbapenem-hydrolyzing beta-lactamase, KPC-1, from a carbapenem-resistant strain of Klebsiella pneumoniae. Antimicrob Agents Chemother. 2001;45(4):1151-61. [PubMed ID: 11257029]. [PubMed Central ID: PMC90438]. https://doi.org/10.1128/AAC.45.4.1151-1161.2001.
-
9.
Pitout JDD, Peirano G, Kock MM, Strydom KA, Matsumura Y. The global ascendency of OXA-48-type carbapenemases. Clin Microbiol Rev. 2019;33(1). [PubMed ID: 31722889]. [PubMed Central ID: PMC6860007]. https://doi.org/10.1128/CMR.00102-19.
-
10.
Maiden MC, Bygraves JA, Feil E, Morelli G, Russell JE, Urwin R, et al. Multilocus sequence typing: A portable approach to the identification of clones within populations of pathogenic microorganisms. Proc Natl Acad Sci U S A. 1998;95(6):3140-5. [PubMed ID: 9501229]. [PubMed Central ID: PMC19708]. https://doi.org/10.1073/pnas.95.6.3140.
-
11.
Diancourt L, Passet V, Verhoef J, Grimont PA, Brisse S. Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. J Clin Microbiol. 2005;43(8):4178-82. [PubMed ID: 16081970]. [PubMed Central ID: PMC1233940]. https://doi.org/10.1128/JCM.43.8.4178-4182.2005.
-
12.
Ibarz Pavon AB, Maiden MC. Multilocus sequence typing. Methods Mol Biol. 2009;551:129-40. [PubMed ID: 19521872]. [PubMed Central ID: PMC3988353]. https://doi.org/10.1007/978-1-60327-999-4_11.
-
13.
Mahon CR, Lehman DC, Manuselis J. Textbook of diagnostic microbiology-e-book. Elsevier Health Sciences; 2018.
-
14.
Wayne P. Performance standards for antimicrobial susceptibility testing M100-S22. Pennsylvania: CLSI; 2018. Available from: http://file.qums.ac.ir/repository/mmrc/CLSI-2018-M100-S28.pdf.
-
15.
Dashti AA, Jadaon MM, Abdulsamad AM, Dashti HM. Heat treatment of bacteria: a simple method of DNA extraction for molecular techniques. Kuwait Med J. 2009;41(2):117-22.
-
16.
Poirel L, Walsh TR, Cuvillier V, Nordmann P. Multiplex PCR for detection of acquired carbapenemase genes. Diagn Microbiol Infect Dis. 2011;70(1):119-23. [PubMed ID: 21398074]. https://doi.org/10.1016/j.diagmicrobio.2010.12.002.
-
17.
Gona F, Comandatore F, Battaglia S, Piazza A, Trovato A, Lorenzin G, et al. Comparison of core-genome MLST, coreSNP and PFGE methods for Klebsiella pneumoniae cluster analysis. Microb Genom. 2020;6(4). [PubMed ID: 32149598]. [PubMed Central ID: PMC7276701]. https://doi.org/10.1099/mgen.0.000347.
-
18.
Papp-Wallace KM, Endimiani A, Taracila MA, Bonomo RA. Carbapenems: past, present, and future. Antimicrob Agents Chemother. 2011;55(11):4943-60. [PubMed ID: 21859938]. [PubMed Central ID: PMC3195018]. https://doi.org/10.1128/AAC.00296-11.
-
19.
Neuner EA, Yeh JY, Hall GS, Sekeres J, Endimiani A, Bonomo RA, et al. Treatment and outcomes in carbapenem-resistant Klebsiella pneumoniae bloodstream infections. Diagn Microbiol Infect Dis. 2011;69(4):357-62. [PubMed ID: 21396529]. [PubMed Central ID: PMC3058153]. https://doi.org/10.1016/j.diagmicrobio.2010.10.013.
-
20.
Rahbar M, Kabeh-Monnavar M, Vatan KK, Fadaei-Haqi A, Shakerian F. Carbapenem resistance in gram-negative bacilli isolates in an Iranian 1000-bed Tertiary Hospital. Pak J Med Sci. 2008;24(4):537-40.
-
21.
Mansouri S, Abbasi S. Prevalence of multiple drug resistant clinical isolates of extended-spectrum beta-lactamase producing Enterobacteriaceae in Southeast Iran. Iran J Med Sci. 2010;35(2):101-8.
-
22.
Mehd SDM, Asghar MS, Kazem SYM, Abdolaziz RL, Zahra R, Sovan AY. [Antimicrobial resistance trends of klebsiella spp. Isolated from patients in Imam Khomeini hospital]. Payavard Salamat. 2012;6(4). Persian.
-
23.
Amin A, Ghumro P, Hussain S, Hameed A. Prevalence of antibiotic resistance among clinical isolates of Kleibsiella pneumoniaeisolated from a Tertiary Care Hospital in Pakistan. Malays J Microbiol. 2009;5(2):81-6. https://doi.org/10.21161/mjm.13409.
-
24.
Ishii Y, Alba J, Kimura S, Shiroto K, Yamaguchi K. Evaluation of antimicrobial activity of β-lactam antibiotics using Etest against clinical isolates from 60 medical centres in Japan. Int J Antimicrob Agents. 2005;(4):296-301. [PubMed ID: 15784308].
-
25.
Al Shara MA. Emerging antimicrobial resistance of klebsiella pneumonia strains isolated from pediatric patients in Jordan. Iraqi J Med. 2011;7(2):29-32.
-
26.
Gurung S, Kafle S, Dhungel B, Adhikari N, Thapa Shrestha U, Adhikari B, et al. Detection of OXA-48 gene in carbapenem-resistant Escherichia coli and klebsiella pneumoniae from urine samples. Infect Drug Resist. 2020;13:2311-21. [PubMed ID: 32765007]. [PubMed Central ID: PMC7369300]. https://doi.org/10.2147/IDR.S259967.
-
27.
Soroush Z, Ghane M. Molecular identification of CTX-M, TEM and SHV β-lactamases in Klebsiella pneumoniae isolated from respiratory system of patients in the ICU of educational hospitals in Tehran. KAUMS J (FEYZ). 2017;21(3):232-9.
-
28.
Khairy RMM, Mahmoud MS, Shady RR, Esmail MAM. Multidrug-resistant Klebsiella pneumoniae in hospital-acquired infections: Concomitant analysis of antimicrobial resistant strains. Int J Clin Pract. 2020;74(4). e13463. [PubMed ID: 31830351]. https://doi.org/10.1111/ijcp.13463.
-
29.
Tsakris A, Kristo I, Poulou A, Themeli-Digalaki K, Ikonomidis A, Petropoulou D, et al. Evaluation of boronic acid disk tests for differentiating KPC-possessing Klebsiella pneumoniae isolates in the clinical laboratory. J Clin Microbiol. 2009;47(2):362-7. [PubMed ID: 19073868]. [PubMed Central ID: PMC2643660]. https://doi.org/10.1128/JCM.01922-08.
-
30.
Gopalakrishnan S, Kamalanathan A, Rajan S, Bhagat VM, Ali MKS. Emergence of armA and rmtB genes among VIM, NDM, and IMP metallo-beta-lactamase-producing multidrug-resistant Gram-negative pathogens. Acta Microbiol Immunol Hung. 2018;65(1):107-18. [PubMed ID: 28870092]. https://doi.org/10.1556/030.64.2017.027.
-
31.
El-Kholy AA, Elanany MG, Sherif MM, Gad MA. High prevalence of VIM, KPC, and NDM expression among surgical site infection pathogens in patients having emergency surgery. Surg Infect (Larchmt). 2018;19(6):629-33. [PubMed ID: 29979638]. https://doi.org/10.1089/sur.2018.088.
-
32.
Adam MA, Elhag WI. Prevalence of metallo-beta-lactamase acquired genes among carbapenems susceptible and resistant Gram-negative clinical isolates using multiplex PCR, Khartoum hospitals, Khartoum Sudan. BMC Infect Dis. 2018;18(1):668. [PubMed ID: 30558551]. [PubMed Central ID: PMC6296134]. https://doi.org/10.1186/s12879-018-3581-z.
-
33.
Peymani A, Nahaei M, Farajnia S, Hasani A, Mirsalehian A, Sohrabi N, et al. High prevalence of metallo-b-lactamase-producing Acinetobacter baumannii in a teaching hospital in Tabriz, Iran. Jpn J Infect Dis. 2011;64(1):69-71.
-
34.
Pavez M, Mamizuka EM, Lincopan N. Early dissemination of KPC-2-producing Klebsiella pneumoniae strains in Brazil. Antimicrob Agents Chemother. 2009;53(6):2702. [PubMed ID: 19332672]. [PubMed Central ID: PMC2687248]. https://doi.org/10.1128/AAC.00089-09.
-
35.
Nordmann P, Naas T, Poirel L. Global spread of Carbapenemase-producing Enterobacteriaceae. Emerg Infect Dis. 2011;17(10):1791-8. [PubMed ID: 22000347]. [PubMed Central ID: PMC3310682]. https://doi.org/10.3201/eid1710.110655.
-
36.
Bratu S, Landman D, Haag R, Recco R, Eramo A, Alam M, et al. Rapid spread of carbapenem-resistant Klebsiella pneumoniae in New York City: a new threat to our antibiotic armamentarium. Arch Intern Med. 2005;165(12):1430-5. [PubMed ID: 15983294]. https://doi.org/10.1001/archinte.165.12.1430.
-
37.
Meng X, Yang J, Duan J, Liu S, Huang X, Wen X, et al. Assessing molecular epidemiology of carbapenem-resistant Klebsiella pneumoniae (CR-KP) with MLST and MALDI-TOF in central China. Sci Rep. 2019;9(1):2271. [PubMed ID: 30783127]. [PubMed Central ID: PMC6381170]. https://doi.org/10.1038/s41598-018-38295-8.
-
38.
Bushnell G, Mitrani-Gold F, Mundy LM. Emergence of New Delhi metallo-beta-lactamase type 1-producing enterobacteriaceae and non-enterobacteriaceae: global case detection and bacterial surveillance. Int J Infect Dis. 2013;17(5):e325-33. [PubMed ID: 23332300]. https://doi.org/10.1016/j.ijid.2012.11.025.
-
39.
Wailan AM, Paterson DL. The spread and acquisition of NDM-1: A multifactorial problem. Expert Rev Anti Infect Ther. 2014;12(1):91-115. [PubMed ID: 24308710]. https://doi.org/10.1586/14787210.2014.856756.
-
40.
Kim YJ, Kim S, Kim J, Bae S. Tracking short-term changes in the genetic diversity and antimicrobial resistance of OXA-232-producing Klebsiella pneumoniae ST14 in clinical settings. Clin Microbiol Infect. 2020;26(1):78-86. [PubMed ID: 31128287]. https://doi.org/10.1016/j.cmi.2019.05.008.
-
41.
Moubareck CA, Mouftah SF, Pal T, Ghazawi A, Halat DH, Nabi A, et al. Clonal emergence of Klebsiella pneumoniae ST14 co-producing OXA-48-type and NDM carbapenemases with high rate of colistin resistance in Dubai, United Arab Emirates. Int J Antimicrob Agents. 2018;52(1):90-5. [PubMed ID: 29530587]. https://doi.org/10.1016/j.ijantimicag.2018.03.003.
-
42.
Arena F, Spanu T, Henrici De Angelis L, Liotti FM, D'Andrea MM, Menchinelli G, et al. First case of bacteremic liver abscess caused by an ST260-related (ST1861), hypervirulent Klebsiella pneumoniae. J Infect. 2016;73(1):88-91. [PubMed ID: 27084307]. https://doi.org/10.1016/j.jinf.2016.04.006.