Abstract
Background:
RNA polymerase beta subunit (rpoB) gene analysis in bacterial communities is known as a method for determining rifampin sensitivity and genetic diversity among Brucella spp. Detection of antibiotic resistance among Brucella isolates can be a critical approach to control brucellosis. However, rpoB gene analysis of Brucella melitensis for assessing rifampicin resistance has not yet been performed in Iran, which is considered an endemic area for brucellosis.Objectives:
The aim of this study was to analyze the whole sequence of rpoB genes of different B. melitensis isolates from humans to identify the single-nucleotide polymorphisms (SNPs) and mutations related to rifampin resistance and to analyze the genetic diversity of these bacteria in Iran.Methods:
Between 2017 and 2019, a total of 156 blood samples along with 12 synovial fluid specimens were collected from brucellosis patients in different Iranian provinces and subjected to bacterial culture in Brucella selective media. Brucella identification was carried out using classical biotyping and molecular examinations. Polymerase chain reaction (PCR)-based amplification of the rpoB gene was performed by specific rpoB primers for whole gene sequencing. The antimicrobial susceptibility of Brucella isolates was assessed using disk diffusion susceptibility tests and minimal inhibitory concentration (MIC) methods. The presence of rifampin-binding sites and SNPs were investigated through rpoB whole gene sequencing.Results:
Clinical B. melitensis isolates were obtained from blood (13) and synovial fluid (1) samples of patients from different regions of Iran. The results of MIC and disk diffusion susceptibility tests showed that all the isolates were sensitive to rifampin except for one isolate showing intermediate rifampin resistance based on the standards defined for slow-growing bacteria by the Clinical and Laboratory Standards Institute (CLSI). Gene analysis for identifying the mutations related to rifampin resistance and investigating genetic diversity showed that none of the B. melitensis isolates had missense mutations, confirming the susceptibility of all the studied isolates to rifampin.Conclusions:
The present study revealed that rpoB gene analysis could be used for the efficient and precise identifying of the mutations related to rifampin resistance, investigating rifampin binding sites, and genotyping Brucella species. Furthermore, the identification of B. melitensis isolates with intermediate resistance to rifampicin highlighted the importance of periodically carrying out antibiotic susceptibility testing. The molecular detection of rpoB mutations in different Brucella isolates may help to prevent the spread of rifampin-resistant Brucella spp. among humans and livestock.Keywords
1. Background
Brucellosis is known as a zoonotic infection caused by Brucella spp. belonging to the class of alpha-proteobacteria and the family of Brucellaceae. This disease is responsible for high morbidity among animals and humans, leading to considerable health and economic costs in many countries (1). The main species responsible for brucellosis in Iran are Brucella melitensis and B. abortus (1). According to different Iranian reports, B. melitensis has been identified as the most prevalent bacteria responsible for human brucellosis; however, B. abortus has also been reported to a much lesser extent (1). On the other hand, antibiotic therapy is the main approach used for the efficient treatment of human brucellosis. Nonetheless, the intracellular properties of this pathogen cause the relapse of the disease in 5 - 14% of the patients subjected to antibiotic treatment (2, 3).
A six-week course of combination therapy with rifampin and doxycycline is recommended as the treatment of choice by the World Health Organization (WHO) (4). Rifampin combined with quinolones appeared to be significantly less efficient compared to the combination of doxycycline with either streptomycin or rifampin. Rifampin is an important and efficient antibiotic for treating brucellosis and has been repeatedly recommended as the best first-line therapy (4, 5). This antibiotic has a good bacteriostatic or bactericidal effect with ideal intracellular penetration and clear synergism with other antibiotics proposed by the WHO for brucellosis treatment (6). However, there have been several reports on increasing rifampin resistance rates among Brucella isolates from Iran (7-9), Egypt (10), Turkey (11), Saudi Arabia (12), and Qatar (13). Moreover, combined rifampin and doxycycline administration poses problems in Middle East countries because of its potential to stimulate rifampin resistance in other infections, especially tuberculosis (2).
Accordingly, antimicrobial susceptibility testing by E-tests as well as SNP analysis have been recommended as efficient approaches to monitor the effectiveness of antibiotics in the treatment of human brucellosis. This is particularly the case when studying B. melitensis clinical populations in endemic regions (14). On the other hand, the use of the DNA-directed RNA polymerase subunit beta (rpoB ) gene, expressing the b-subunit of RNA polymerase, appeared to be highly effective for detecting the rpoB mutations causing rifampin resistance (15). The evaluation of the rifampin susceptibility profiles of B. melitensis and B. abortus is critical as this antibiotic is the most widely recommended and applied therapeutic agent for the treatment of human brucellosis (16, 17).
Resistance to rifampin is a growing problem in both developed and developing countries (18-20). This issue is even more difficult to handle in patients with immunodeficiency disorders (14, 21). Therefore, the rapid detection of antibiotic resistance is of overwhelming importance for the efficient management of brucellosis. Some investigations put into light the genetic basis of resistance against anti-brucellosis antibiotics among Brucella isolates (15, 22). Resistance against rifampin is most possibly due to point mutations as well as small insertions and deletions in the rpoB gene expressing the RNA polymerase beta subunit (23, 24). Also, some mutation sites in the rpoB gene have been identified as biomarkers for rifampin resistance in Brucella spp. (25).
Moreover, rpoB genotyping allows the identification of new bacterial species and facilitates the analysis of bacterial communities (26, 27). The frequency and nature of rpoB gene mutations vary significantly among different bacterial species (23, 24). However, to our knowledge, there is no data available on specific mutational patterns of rifampin resistance in the Brucella strains isolated from Iranian patients. In this study, rpoB mutations of the Brucella strains isolated from different parts of Iran were evaluated by whole rpoB gene sequencing of a 4134-bp segment. We also analyzed specific interactions between the ligands and proteins expressed by rpoB in the isolated Brucella species.
2. Objectives
The aim of the current study was to determine SNP(s) in the rpoB gene by sequencing the whole gene from different clinical isolates of B. melitensis to investigate any association between SNPs and rifampin resistance.
3. Methods
3.1. Bacteria Isolation
A total of 156 blood and 12 synovial fluid specimens were collected from brucellosis patients from different provinces of Iran (Alborz, Qom, Kerman, Tehran, Hamadan, Khorasan Razavi, Kermanshah, and Yazd) and cultured in Brucella selective media. All the samples were collected from 2017 to 2019. Inclusion criteria for patients were positive serological tests and presenting brucellosis symptoms. Patients with positive serological results for the Rose Bengal test (RBT), serum agglutination test (SAT), and 2-Mercaptoethanol (2-ME) test were selected for bacterial culture. The most common symptoms observed among patients were fever and fatigue accompanied with one or more of the following symptoms: night sweats, chills, weight loss, headache, arthralgia, myalgia, weakness, arthritis, and malaise.
All the samples were inoculated into Brucella selective agar supplemented with vancomycin (10.0 mg), cycloheximide (50.0 mg), nystatin (50,000 IU), polymyxin B (2,500 IU), nalidixic acid (2.5 mg), and bacitracin (12,500 IU) (Oxoid, UK) along with 5% inactivated horse serum. Then the culture plates were incubated at 37°C under 10% CO2 for 10 days. The characterization of Brucella isolates was done by classical biotyping according to previous works (1). Genomic bacterial DNA was extracted using fresh cultures by the Exgene Cell SV kit (Gene All, South Korea) based on the manufacturer’s protocol. The DNA integrity of the isolated bacteria was evaluated by 1.5 % agarose gel electrophoresis, and DNA concentration was evaluated by reading ODs at 260/280 nm by a Nanodrop Spectrophotometer. Bacterial DNA was stored at -20°C until further analysis.
3.2. Molecular Typing
The extracted genomic DNA was subjected to species-level identification according to IS711-based PCR (AMOS-PCR) and Bruce-ladder PCR to detect Brucella spp. The AMOS-PCR amplification with 5-primer multiplex was done at the thermal program of one cycle of denaturation at 95°C (5 min) and 40 cycles of denaturation at 95°C (30 s), annealing at 55°C (60 s), and extension at 72°C (3 min), followed by a final extension at 72°C (10 min) (28). Molecular typing by multiplex PCR (Bruce-ladder) with 16-primer multiplex PCR was done applying an initial denaturation at 95°C (5 min) and 35 cycles of denaturation at 95°C (30 s), annealing at 56°C (90 s), and extension at 72°C (3 min), followed by a final extension at 72°C (10 min) (29). Brucella abortus 544 and B. melitensis 16M were used as appropriate positive controls for PCR experiments. A sample without DNA was used as the negative control. The amplicons were separated by 1.5 % agarose gel electrophoresis. All the PCR primers used have been shown in Table 1.
Primer Sets and Expected Amplicon Sizes Related to Different Brucella Species
| Strain Amplicon & Primer Set | Primer Sequence (5’-3’) | DNA Target | Size (bp) | References |
|---|---|---|---|---|
| AMOS PCR | IS711 | 498 | (28) | |
| IS711f | TGCCGATCACTTTCAAGGGCCTTCAT | |||
| AB r | GACGAACGGAATTTTTCCAATCCC | |||
| AMOS PCR | IS711 | 731 | (28) | |
| IS711 f | TGCCGATCACTTTCAAGGGCCTTCAT | |||
| BM r | AAATCGCGTCCTTGCTGGTCTGA | |||
| AMOS PCR | IS711 | 976 | (28) | |
| IS711 | TGCCGATCACTTTCAAGGGCCTTCAT | |||
| B. ovis | CGGGTTCTGGCACCATCGTCG | |||
| AMOS PCR | IS711 | 285 | (28) | |
| IS711 | TGCCGATCACTTTCAAGGGCCTTCAT | |||
| B. suis | GCGCGGTTTTCTGAAGGTTCAGG | |||
| Bruce-ladder PCR | Glycosyl transferase, gene wboA | 1,682 | (29) | |
| BMEI0998f | ATC CTA TTG CCC CGATAA GG | |||
| BMEI0997r | GCT TCG CAT TTT CACTGT AGC | |||
| Bruce-ladder PCR | Immunodominant antigen, gene bp26 | 450 | (29) | |
| BMEI0535f | GCG CAT TCT TCG GTTATG AA | |||
| BMEI0536r | CGC AGG CGA AAA CAGCTA TAA | |||
| Bruce-ladder PCR | Outer membrane Protein, gene omp31 | 1071 | (29) | |
| BMEII0843f | TTT ACA CAG GCA ATCCAG CA | |||
| BMEII0844r | GCG TCC AGT TGT TGTTGA TG | |||
| Bruce-ladder PCR | Polysaccharide deacetylase | 794 | (29) | |
| BMEI1436f | ACG CAG ACG ACC TTCGGTAT | |||
| BMEI1435r | TTT ATC CAT CGC CCTGTCAC | |||
| Bruce-ladder PCR | Erythritol catabolism, gene eryC (Derythrulose- 1-phosphate dehydrogenase) | 587 | (29) | |
| BMEII0428f | GCC GCT ATT ATG TGGACT GG | |||
| BMEII0428r | AAT GAC TTC ACG GTCGTT CG | |||
| Bruce-ladder PCR | ABC transporter binding protein | 272 | (29) | |
| BR0953f | GGA ACA CTA CGC CACCTT GT | |||
| BR0953r | GAT GGA GCA AAC GCTGAA G | |||
| Bruce-ladder PCR | Ribosomal protein S12, gene rpsL | 218 | (29) | |
| BMEI0752f | CAG GCA AAC CCT CAG AAG C | |||
| BMEI0752r | GAT GTG GTA ACG CAC ACC AA | |||
| Bruce-ladder PCR | Transcriptionalregulator, CRP family | 152 | (29) | |
| BMEII0987f | CGC AGA CAG TGA CCATCA AA | |||
| BMEII0987r | GTA TTC AGC CCC CGTTAC CT | |||
| rpoB | rpoB gene | 4134 | (15, 25) | |
| 1rB | ATGGCTCAGACCCATTCTTTC | |||
| 4134rB | TTATTCTGCCGCGTCCGGAA |
3.3. RpoB Gene Amplification
The PCR amplification of the rpoB gene was performed using specific rpoB primers for whole gene amplification (15). The reaction mixture (25 μL ) consisted of 0.5 mM of each primer, 0.05 IU of Taq polymerase, 0.2 mM deoxynucleotide triphosphate, 10 mM Tris–HCl (pH 8), 50 mM KCl, 1.5 mM MgCl2, and 100 ng genomic DNA (evaluated by reading O.D. at 260 nm) (27, 30).
3.4. Sequence Analysis of rpoB Amplicons
After purification by a PCR purification kit, an ABI Prism 377 sequencer Genetic Analyzer (Applied Biosystems Foster City, CA) was utilized for sequencing PCR products based on the Big-Dye Terminator protocol (Applied Biosystems) (Gene AlI, Seoul, South Korea). The assembling of rpoB sequences was performed by Mega 6 software, and the amino acids were deduced according to the CLUSTAL W (31). For nucleotide diversity evaluation, all rpoB sequences were generated by specific primers and compared to those available in the NCBI database. Mutations were analyzed twice to confirm our findings.
3.5. Antimicrobial Susceptibility Testing
The minimum inhibitory concentration (MIC) of rifampin against clinical Brucella isolates was determined using the disk-diffusion susceptibility test (5 μg rifampin per disk) and rifampin E-test (0.016 - 256 μg/mL) (E-test®, Liofilchem, Italy) following CLSI guidelines. Bacterial suspensions were prepared for each isolate from fresh and pure colonies with 0.5 McFarland turbidity standards and spread onto Muller-Hinton agar (Sigma- Aldrich) supplemented with 5% sheep’s blood. All the isolates were incubated at 37°C with 10% CO2, and the results were read after 48 hours. The breakpoints of Brucella isolates against rifampin were presented as MIC50 and MIC90. The CLSI guideline for slow-growing bacteria (Haemophilus spp.) was used for interpreting the results (32). All antibiotics were assessed in duplicate against all the isolates. Furthermore, Escherichia coli (ATCC 25922) and B. melitensis 16M strain (ATCC 23456) were applied as reference strains in antimicrobial susceptibility tests.
3.6. Rifampin-Binding Site and SNP Analyses for the rpoB Gene
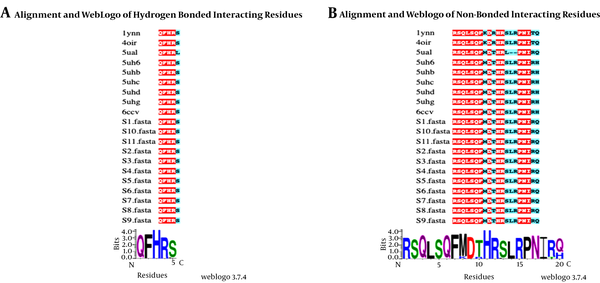
Nine crystal structures (Thermus acquaticus (1YNN), T. thermophilus (4OIR), Mycobacterium smegmatis (6CCV), E. coli (5UAL), and M. tuberculosis (5UHC, 5UHB, 5UHD, 5UHG, and 5UH6)) bound to rifampin, and related molecules were identified by searching the PDB database. An in-house python script was written, performing multiple sequence alignments to compare the binding sites of the provided crystal structures with 11 clinical isolates. The generated alignment for the binding site was color-coded, allowing for discerning identical and mutated positions in the sequence. Furthermore, the mutations related to rifampin resistance were investigated at Cd 154 (GTT/TTT), Cd 526 (GAC/TAC, GAC/AAC, GAC/GGC), Cd 536 (CAC/CTC, CAC/TAC), Cd 539 (CGC/AGC), Cd 541 (TCG/TTG), and Cd 574 (CCG/CTG) of the rpoB gene by aligning with the reference sequence using Mega 6 software (22).
4. Results
4.1. Bacteria Isolation and Identification
Typical phenotypic features of Brucella spp., such as small smooth-surface honey-colored and translucent, shiny colonies, were observed in 14 isolated bacteria. A collection of clinical B. melitensis isolates (blood (13), synovial fluid (1)) was obtained from different regions of Iran. All the isolated bacteria grew after five days of incubation at 37°C under 10% carbon dioxide (CO2). The isolated bacteria were Gram-negative with biovar levels as B. melitensis (i.e., biovar one or three).
4.2. Molecular Identification
The identity of Brucella species was confirmed at the molecular level using AMOS-PCR and Bruce-ladder PCR. A total of 11 B. melitensis strains were isolated from 168 human samples. These strains belonged to two different biovars, and the biovar one (10 cases from Qom, Kerman, Tehran, Hamadan, Khorasan Razavi, Kermanshah, and Yazd provinces) was more common than the biovar three (4 cases from Alborz province). All the isolated bacteria were recognized as wild type B. melitensis by both AMOS-PCR (a PCR product of 731 bp, Figure 1) and Bruce-ladder PCR (products of 1682, 794, 587, 450, 152, and 1,071 bp, Figure 2).
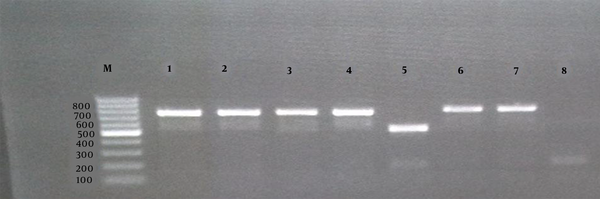
Agarose gel electrophoresis (1%) of the PCR products amplified by the AMOS-PCR of bacterial DNA samples. The “M” shows the DNA marker (100 bp DNA ladder). Lanes 1, 2, 3, 4, and 7 show Brucella melitensis field strains; Lane 5, B. abortus 544 as the reference; Lane 6, B. melitensis 16 M as the reference; Lane 8, negative control.
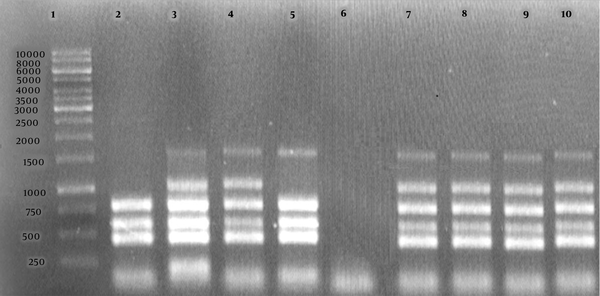
Agarose gel electrophoresis (1%) of the PCR products amplified by the Bruce-ladder PCR of bacterial DNA samples. Lane 1 shows the DNA marker (1000 bp DNA ladder). Lane 2 shows Brucella abortus RB51; Lane 3, B. melitensis Rev1; Lane 4, B. melitensis 16 M; Lane 5, B. abortus 544; Lane 6, negative control, Lanes 7 - 10, B. melitensis field strains.
4.3. Analysis of Genetic Distances
The rpoB gene of the 14 clinical Brucella isolates was characterized through full-length sequencing. The nucleotide and deduced amino acid sequences of Brucella isolates were edited by Mega 6 software and deposited in the NCBI database with the accession numbers of MK790247, MK598748, MK790249, MK790248, MK790251, MK790252, MK629658, MK629659, MK629660, MK790250, MK629661, MW589198, MW168443, and MW589199. All nucleotides and deduced amino acids were scrutinized to detect missense mutations. Nucleotide sequences of the rpoB gene among the Brucella isolates showed 99% identity. The results obtained for reference strains, including B. melitensis B115, Rev 1, 16M, and B. melitensis, along with other isolates identified from other regions, were compared with those resulting from this study. Gene analysis for identifying the mutations related to rifampin resistance and evaluating genetic diversity showed that none of the B. melitensis isolates had missense mutations, confirming the fact that all the clinical isolates from different provinces were susceptible to rifampin. Also, according to our results, rpoB typing grouped the majority of the isolates from provinces of Alborz, Qom, Kerman, Hamadan, Khorasan Razavi, Kermanshah, and Yazd in the rpoB type 2 (629-Val (GTG), 985-Val (GTC), 1249-Met (ATG), and 1309-Leu (CTA)) while only one of them (from Tehran) belonged to the rpoB type 1 (629-Ala (GCG), 985-Ala (GCC), 1249-Met (ATG) and 1309-Leu (CTG)) (Table 2).
The Results of the Disk Diffusion Test and E-test Compared with the Sequencing Results for Mutation-Specific Codons Related to Rifampicin Resistance in Brucella melitensis
| Brucella melitensis Isolates a | Isolates' Properties | Mutation Specific Codons Associated with Rifampicin-Resistant Phenotype b | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Accession numbers | Province | Isolates | Genotyp | MIC resultsc (µg/ml) | Disk diffusion d (mm) | Cde 154GTT | Cd 526 GAC | Cd 536 CAC | Cd 539 CGC | Cd 541 TCG | Cd 574 CCG | |
| 16 M | Reference strain | - | Reference strain | 2 | 0.75 | 38 | GTT | GAC | CAC | CGC | TCG | CCG |
| B. melitensis bv3 | MK629658 | Alborz | Blood | 2 | 1 | 22 | GTT | GAC | CAC | CGC | TCG | CCG |
| B. melitensis bv3 | MK629659 | Alborz | Blood | 2 | 0.38 | 29 | GTT | GAC | CAC | CGC | TCG | CCG |
| B. melitensis bv1 | MK629660 | Alborz | Blood | 2 | 2 | 17 | GTT | GAC | CAC | CGC | TCG | CCG |
| B. melitensis bv1 | MK629661 | Qom | Blood | 2 | 1 | 22 | GTT | GAC | CAC | CGC | TCG | CCG |
| B. melitensis bv1 | MK790247 | Kerman | Blood | 2 | 0.38 | 30 | GTT | GAC | CAC | CGC | TCG | CCG |
| B. melitensis bv1 | MK790248 | Tehran | Synovial fluid | 1 | 0.38 | 28 | GTT | GAC | CAC | CGC | TCG | CCG |
| B. melitensis bv1 | MK598748 | Alborz | Blood | 2 | 0.38 | 29 | GTT | GAC | CAC | CGC | TCG | CCG |
| B. melitensis bv1 | MK790249 | Khorasan | Blood | 2 | 0.38 | 27 | GTT | GAC | CAC | CGC | TCG | CCG |
| B. melitensis bv1 | MK790250 | Hamadan | Blood | 2 | 0.38 | 30 | GTT | GAC | CAC | CGC | TCG | CCG |
| B. melitensis bv1 | MK790251 | Kermanshah | Blood | 2 | 1 | 22 | GTT | GAC | CAC | CGC | TCG | CCG |
| B. melitensis bv1 | MK790252 | Yazd | Blood | 2 | 0.38 | 33 | GTT | GAC | CAC | CGC | TCG | CCG |
| B. melitensis bv3 | MW589198 | Alborz | Blood | 2 | 0.38 | 35 | GTT | GAC | CAC | CGC | TCG | CCG |
| B. melitensis bv1 | MW168443 | Alborz | Blood | 2 | 0.125 | 38 | GTT | GAC | CAC | CGC | TCG | CCG |
| B. melitensis bv3 | MW589199 | Alborz | Blood | 2 | 1 | 25 | GTT | GAC | CAC | CGC | TCG | CCG |
4.4. Antimicrobial Susceptibility Testing
According to CLSI breakpoints for slow-growing bacteria (Haemophilus spp.), we evaluated the antibiotic sensitivity of the collected isolates. Considering the antibiotic sensitivity of the isolated strains based on the disk diffusion test, inhibition zone diameters ranged from 17 to 38 mm. The range of MIC values for rifampin was from 0.125 to 2 μg/mL. All the clinical isolates were sensitive to rifampin except for one of them, which revealed intermediate rifampin resistance. The MIC50 and MIC90 values were reported as 0.38 and 1 µg/ml, respectively. The intermediate rifampin resistance observed in one of the isolates could probably lead to resistant phenotype (Table 2).
4.5. RpoB Analysis for Detecting SNPs
The rifampin-resistant strain showed no mutations at the codons 154, 526, 536, 539, 541, and 574 of the rpoB gene (Figure 3). Eleven rpoB sequences were aligned with nine rpoB crystal structures from T.s acquaticus (1YNN), T. thermophilus (4OIR), M. smegmatis (6CCV), E. coli (5UAL), and M. tuberculosis (5UHC, 5UHB, 5UHD, 5UHG, and 5UH6). In the present study, the rpoB sequence identity with the sequences of their crystal structures ranged from 53% to 57%. Furthermore, binding residues were mapped to analyze conservation in the rpoB rifampin-binding site among the isolates through the comparison of the identified binding residues with structure-ligand complexes (Figure 3).
A, The amino acids forming hydrogen bonds with rifampin in PDB structures (identified from the PDB sum database). B, The amino acids that formed non-bonded interactions with rifampin. Identical amino acid positions have been shaded in red whereas cyan-shaded positions indicate amino acid substitutions. A web logo has been presented for both bonded and non-bonded interactions.
5. Discussion
The worldwide distribution of Brucella spp. infections in different hosts has highlighted the crucial need for various local laboratories to apply the same typing methods to facilitate data exchange and comparison. According to our results, rpoB typing showed that most of the assessed Iranian Brucella isolates belonged to the rpoB type 2. These findings are in agreement with those of a study showing similar missense mutations at the same location (i.e., the codon 985) of the rpoB gene in three B. melitensis type 2 strains (25, 27). Other rpoB missense mutations of the genotype 2 of B. melitensis have also been observed in other studies (25-27, 30). The genotype 1 of B. melitensis, harboring the rpoB gene type 1, was identified in a single isolate from Iranian capital, Tehran (accession number: MK790248). On the other side, rpoB type 2 was found in all the other isolates collected from other Iranian provinces.
In this study, we combined microbiological and genomic analyses to evaluate specific ligand-protein interactions with rifampin according to the rpoB gene of Brucella spp. For this purpose, 14 Brucella isolates were recovered from 168 human specimens. We sequenced the full-length rpoB gene and investigated the discriminative power of rifampin interactive properties. The rpoB is a highly conserved housekeeping gene that is present in all bacteria due to its critical function in cellular metabolism (25, 26). Indeed, amino acid substitutions secondary to rpoB point mutations have been demonstrated to have important implications in the development of rifampin resistance. In previous studies, two B. melitensis strains showed the following mutations: Val154Phe, Asp526Tyr/Asn/Gly, His536Leu/Tyr, Arg539Ser, Ser541Leu, and Pro574Leu, leading to rifampin resistance (15).
Rifampin is one of the broad-spectrum and most effective antibiotics used in multidrug regimens to treat brucellosis. Resistance against rifampin in Brucella spp. can easily occur in human infections (15). It has been reported that rpoB gene mutations can be involved in the development of rifampin resistance in different bacterial strains such as Brucella species (25, 26). In our analysis, all the B. melitensis isolated were recognized as rifampin-susceptible based on molecular testing. These findings were further confirmed by the results of the E-test, as well as disk-diffusion susceptibility tests. We found no reports investigating the molecular mechanisms of rifampin-resistance among B. melitensis bacteria in Iranian patients with brucellosis. In a recent investigation in our institution, in vitro antimicrobial susceptibility tests showed a MIC90 value of 1.5 mg/L and up to 1.7% intermediate susceptibility to rifampin in B. melitensis isolates (7). However, our results showed that all the isolates tested in this study were susceptible to rifampin. Moreover, none of them showed the most frequently observed change (i.e., His 536) in B. melitensis isolates. All the isolates displayed a common sequence of rpoB at the position of 125 to 720 (a fragment with 595 amino acids), which is specific for B. melitensis 16M. Our results confirmed the findings of previous studies showing neither mutations nor rifampin resistance in isolates with intermediate susceptibility to rifampin (22, 30).
The present rpoB data, as well as the characterization of rpoB mutations, showed that the B. melitensis isolates originating from the patients first diagnosed with brucellosis were all susceptible to rifampin. Various investigations on Iranian samples have also reported similar susceptibility findings based on the MIC method (MIC50 and 90 as 0.38 and 1 µg/mL, MIC range of 0.125 - 1.5 mg/L) (7, 33). Therefore, the selection of rifampin as the first choice for the treatment of brucellosis is supported by these findings. The results of this study also supported the applicability of molecular testing for investigating rifampin susceptibility. On the other hand, in vitro antimicrobial susceptibility testing of Brucella can be performed by several other techniques, including agar dilution, broth microdilution, and the E-test (34). However, antimicrobial susceptibility testing for highly zoonotic bacteria such as Brucella needs a laboratory with level-3 biosafety, which may not be accessible in many regions (35).
Moreover, traditional antibiotic susceptibility tests require several passaging of Brucella, which is a high-risk and time-consuming activity (22). Furthermore, in vitro antimicrobial susceptibility methods for Brucella spp. need proper standardization (36). Therefore, molecular approaches could be safer and more convenient for studying antimicrobial resistance in Brucella spp. Different studies have reported many missense mutations in the rpoB gene of M. tuberculosis, Legionella pneumophila, Helicobacter pylori, and E. coli, developing resistance to rifampicin (37, 38). To date, the susceptibility of B. melitensis clinical isolates to rifampin had not been assessed in Iran using molecular methods based on rpoB gene mutation analysis. In this study; however, no rifampin resistance-associated missense mutations were detected in the rpoB gene of the B. melitensis strains isolated from Iranian patients. Nevertheless, phenotypically intermediate resistance to rifampin is, per se, a matter of concern. Therefore, SNP analysis can be proposed as a useful technique for regular screening of rifampin resistance in B. melitensis isolates.
5.1. Conclusions
According to our results, rpoB gene SNP analysis was performed to characterize rifampin resistance-associated missense mutations in the B. melitensis strains isolated from the Iranian patients first-diagnosed with brucellosis. All the isolates showed a common sequence at the analyzed positions (i.e., Cd 154, Cd 526, Cd 536, Cd 539, Cd 541, and Cd 574) of the rpoB gene, and none of them showed the rpoB gene missense mutations previously reported in rifampicin-resistant B. melitensis. All the studied isolates were found to be rifampicin-susceptible according to molecular analysis. These findings were further confirmed by the results of the E-test and disk-diffusion susceptibility tests. Furthermore, the rpoB gene displayed potential polymorphisms that could be used for the differentiation of all species of Brucella and their biovars. These observations highlight the need for regular monitoring of rifampin susceptibility in Brucella spp. by molecular methods in vitro.
Acknowledgements
References
-
1.
Dadar M, Alamian S, Behrozikhah AM, Yazdani F, Kalantari A, Etemadi A, et al. Molecular identification of Brucella species and biovars associated with animal and human infection in Iran. Vet Res Forum. 2019;10(4):315-21. [PubMed ID: 32206227]. [PubMed Central ID: PMC7065579]. https://doi.org/10.30466/vrf.2018.89680.2171.
-
2.
Solis Garcia del Pozo J, Solera J. Systematic review and meta-analysis of randomized clinical trials in the treatment of human brucellosis. PLoS One. 2012;7(2). e32090. [PubMed ID: 22393379]. [PubMed Central ID: PMC3290537]. https://doi.org/10.1371/journal.pone.0032090.
-
3.
Skalsky K, Yahav D, Bishara J, Pitlik S, Leibovici L, Paul M. Treatment of human brucellosis: Systematic review and meta-analysis of randomised controlled trials. BMJ. 2008;336(7646):701-4. [PubMed ID: 18321957]. [PubMed Central ID: PMC2276295]. https://doi.org/10.1136/bmj.39497.500903.25.
-
4.
Ariza J, Gudiol F, Pallares R, Viladrich PF, Rufi G, Corredoira J, et al. Treatment of human brucellosis with doxycycline plus rifampin or doxycycline plus streptomycin. A randomized, double-blind study. Ann Intern Med. 1992;117(1):25-30. [PubMed ID: 1596044]. https://doi.org/10.7326/0003-4819-117-1-25.
-
5.
Taghvaee MR, Nozadi MS, Hassani M. A comparison between doxycycline-rifampin and ciprofloxacin-rifampin regimens in the treatment of acute Brucellosis. Indian J Med Sci. 2011;65(10):436-43. [PubMed ID: 23511044]. https://doi.org/10.4103/0019-5359.109263.
-
6.
Sandalakis V, Psaroulaki A, De Bock PJ, Christidou A, Gevaert K, Tsiotis G, et al. Investigation of rifampicin resistance mechanisms in Brucella abortus using MS-driven comparative proteomics. J Proteome Res. 2012;11(4):2374-85. [PubMed ID: 22360387]. https://doi.org/10.1021/pr201122w.
-
7.
Alamian S, Dadar M, Etemadi A, Afshar D, Alamian MM. Antimicrobial susceptibility of Brucella spp. isolated from Iranian patients during 2016 to 2018. Iran J Microbiol. 2019;11(5):363-7. [PubMed ID: 32148665]. [PubMed Central ID: PMC7049315].
-
8.
Razzaghi R, Rastegar R, Momen-Heravi M, Erami M, Nazeri M. Antimicrobial susceptibility testing of Brucella melitensis isolated from patients with acute brucellosis in a centre of Iran. Indian J Med Microbiol. 2016;34(3):342-5. [PubMed ID: 27514957]. https://doi.org/10.4103/0255-0857.188336.
-
9.
Farazi A, Hoseini SD, Ghaznavirad E, Sadekhoo S. Antibiotic susceptibility of brucella melitensis in markazi province of Iran. Int J Infect Dis. 2018;73. https://doi.org/10.1016/j.ijid.2018.04.3697.
-
10.
Abdel-Maksoud M, House B, Wasfy M, Abdel-Rahman B, Pimentel G, Roushdy G, et al. In vitro antibiotic susceptibility testing of Brucella isolates from Egypt between 1999 and 2007 and evidence of probable rifampin resistance. Ann Clin Microbiol Antimicrob. 2012;11(1):14. [PubMed ID: 22929054]. [PubMed Central ID: PMC3464789]. https://doi.org/10.1186/1476-0711-11-24.
-
11.
Etiz P, Kibar F, Ekenoğlu Y, Yaman A. Characterization of antibiotic susceptibility of brucella spp isolates with E-Test method. Arch Clin Microbiol. 2015;6.
-
12.
Shaalan MA, Memish ZA, Mahmoud SA, Alomari A, Khan MY, Almuneef M, et al. Brucellosis in children: Clinical observations in 115 cases. Int J Infect Dis. 2002;6(3):182-6. https://doi.org/10.1016/s1201-9712(02)90108-6.
-
13.
Deshmukh A, Hagen F, Sharabasi OA, Abraham M, Wilson G, Doiphode S, et al. In vitro antimicrobial susceptibility testing of human Brucella melitensis isolates from Qatar between 2014 - 2015. BMC Microbiol. 2015;15(1):121. [PubMed ID: 26073177]. [PubMed Central ID: PMC4466872]. https://doi.org/10.1186/s12866-015-0458-9.
-
14.
Villate SCA, Casallas JCG. Update of antibiotic therapy of brucellosis. New insight into brucella infection and foodborne diseases. IntechOpen; 2020.
-
15.
Marianelli C, Ciuchini F, Tarantino M, Pasquali P, Adone R. Genetic bases of the rifampin resistance phenotype in Brucella spp. J Clin Microbiol. 2004;42(12):5439-43. [PubMed ID: 15583262]. [PubMed Central ID: PMC535235]. https://doi.org/10.1128/JCM.42.12.5439-5443.2004.
-
16.
Ariza J, Bosilkovski M, Cascio A, Colmenero JD, Corbel MJ, Falagas ME, et al. Perspectives for the treatment of brucellosis in the 21st century: the Ioannina recommendations. PLoS Med. 2007;4(12). e317. [PubMed ID: 18162038]. [PubMed Central ID: PMC2222927]. https://doi.org/10.1371/journal.pmed.0040317.
-
17.
Tan KK, Tan YC, Chang LY, Lee KW, Nor'e SS, Yee WY, et al. Geographical distribution of Brucella melitensis inferred from rpoB gene variation. J Infect Dev Ctries. 2017;11(5):420-5. [PubMed ID: 30943180]. https://doi.org/10.3855/jidc.7598.
-
18.
Probert WS, Schrader KN, Khuong NY, Bystrom SL, Graves MH. Real-time multiplex PCR assay for detection of Brucella spp., B. abortus, and B. melitensis. J Clin Microbiol. 2004;42(3):1290-3. [PubMed ID: 15004098]. [PubMed Central ID: PMC356861]. https://doi.org/10.1128/JCM.42.3.1290-1293.2004.
-
19.
Ravanel N, Gestin B, Maurin M. In vitro selection of fluoroquinolone resistance in Brucella melitensis. Int J Antimicrob Agents. 2009;34(1):76-81. [PubMed ID: 19261448]. https://doi.org/10.1016/j.ijantimicag.2009.01.002.
-
20.
Dadar M, Tiwari R, Sharun K, Dhama K. Importance of brucellosis control programs of livestock on the improvement of one health. Vet Q. 2021;41(1):137-51. [PubMed ID: 33618618]. [PubMed Central ID: PMC7946044]. https://doi.org/10.1080/01652176.2021.1894501.
-
21.
Jacob NR, Rodriguez CG, Binaghi MA, Scapellato PG, Rosales Ostriz MB, Ayala SM, et al. Brucellosis complicating chronic non-infectious disorders: diagnostic and therapeutic dilemmas. J Med Microbiol. 2008;57(Pt 9):1161-6. [PubMed ID: 18719189]. https://doi.org/10.1099/jmm.0.2008/000687-0.
-
22.
Sayan M, Yumuk Z, Dundar D, Bilenoglu O, Erdenlig S, Yasar E, et al. Rifampicin resistance phenotyping of Brucella melitensis by rpoB gene analysis in clinical isolates. J Chemother. 2008;20(4):431-5. [PubMed ID: 18676221]. https://doi.org/10.1179/joc.2008.20.4.431.
-
23.
Huitric E, Werngren J, Jureen P, Hoffner S. Resistance levels and rpoB gene mutations among in vitro-selected rifampin-resistant Mycobacterium tuberculosis mutants. Antimicrob Agents Chemother. 2006;50(8):2860-2. [PubMed ID: 16870787]. [PubMed Central ID: PMC1538691]. https://doi.org/10.1128/AAC.00303-06.
-
24.
Sharma M, Sethi S, Mishra B, Sengupta C, Sharma SK. Rapid detection of mutations in rpoB gene of rifampicin resistant Mycobacterium tuberculosis strains by line probe assay. Indian J Med Res. 2003;117:76-80. [PubMed ID: 12931842].
-
25.
Marianelli C, Ciuchini F, Tarantino M, Pasquali P, Adone R. Molecular characterization of the rpoB gene in Brucella species: new potential molecular markers for genotyping. Microbes Infect. 2006;8(3):860-5. [PubMed ID: 16483820]. https://doi.org/10.1016/j.micinf.2005.10.008.
-
26.
Sayan M, Yumuk Z, Bilenoglu O, Erdenlig S, Willke A. Genotyping of Brucella melitensis by rpoB gene analysis and re-evaluation of conventional serotyping method. Jpn J Infect Dis. 2009;62(2):160-3. [PubMed ID: 19305062].
-
27.
Bazrgari N, Garosi GA, Dadar M. Genetic diversity and phylogenetic relationship of clinical isolates of brucella melitensis based on gene polymorphism of β subunit of RNA Polymerase (rpoB) gene in Iran. Iran J Med Microbiol. 2020;14(5):425-40. https://doi.org/10.30699/ijmm.14.5.425.
-
28.
Ewalt DR, Bricker BJ. Validation of the abbreviated Brucella AMOS PCR as a rapid screening method for differentiation of Brucella abortus field strain isolates and the vaccine strains, 19 and RB51. J Clin Microbiol. 2000;38(8):3085-6. [PubMed ID: 10921983]. [PubMed Central ID: PMC87192]. https://doi.org/10.1128/JCM.38.8.3085-3086.2000.
-
29.
Lopez-Goni I, Garcia-Yoldi D, Marin CM, de Miguel MJ, Munoz PM, Blasco JM, et al. Evaluation of a multiplex PCR assay (Bruce-ladder) for molecular typing of all Brucella species, including the vaccine strains. J Clin Microbiol. 2008;46(10):3484-7. [PubMed ID: 18716225]. [PubMed Central ID: PMC2566117]. https://doi.org/10.1128/JCM.00837-08.
-
30.
Valdezate S, Navarro A, Villalon P, Carrasco G, Saez-Nieto JA. Epidemiological and phylogenetic analysis of Spanish human Brucella melitensis strains by multiple-locus variable-number tandem-repeat typing, hypervariable octameric oligonucleotide fingerprinting, and rpoB typing. J Clin Microbiol. 2010;48(8):2734-40. [PubMed ID: 20554816]. [PubMed Central ID: PMC2916618]. https://doi.org/10.1128/JCM.00533-10.
-
31.
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30(12):2725-9. [PubMed ID: 24132122]. [PubMed Central ID: PMC3840312]. https://doi.org/10.1093/molbev/mst197.
-
32.
Wayne P. Performance standards for antimicrobial susceptibility testing: Twenty-fourth informational supplement, M100-S24. Clinical and Laboratory Standards Institute (CLSI). 2014;34(1).
-
33.
Asadi FT, Hashemi SH, Alikhani MY, Moghimbeigi A, Naseri Z. Clinical and diagnostic aspects of brucellosis and antimicrobial susceptibility of Brucella isolates in Hamedan, Iran. Jpn J Infect Dis. 2016:JJID. 2016.133.
-
34.
Trujillano-Martìn I, Garcìa-Sánchez E, Fresnadillo MJ, Garcìa-Sánchez JE, Garcìa-Rodrìguez JÁ, Montes Martìnez I. In vitro activities of five new antimicrobial agents against Brucella melitensis. Int J Antimicrob Agents. 1999;12(2):185-6. https://doi.org/10.1016/s0924-8579(99)00049-7.
-
35.
Ackelsberg J, Liddicoat A, Burke T, Szymczak WA, Levi MH, Ostrowsky B, et al. Brucella exposure risk events in 10 clinical laboratories, New York city, USA, 2015 to 2017. J Clin Microbiol. 2020;58(2). [PubMed ID: 31694974]. [PubMed Central ID: PMC6989065]. https://doi.org/10.1128/JCM.01096-19.
-
36.
Orhan G, Bayram A, Zer Y, Balci I. Synergy tests by E test and checkerboard methods of antimicrobial combinations against Brucella melitensis. J Clin Microbiol. 2005;43(1):140-3. [PubMed ID: 15634962]. [PubMed Central ID: PMC540140]. https://doi.org/10.1128/JCM.43.1.140-143.2005.
-
37.
Mariam DH, Mengistu Y, Hoffner SE, Andersson DI. Effect of rpoB mutations conferring rifampin resistance on fitness of Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2004;48(4):1289-94. [PubMed ID: 15047531]. [PubMed Central ID: PMC375340]. https://doi.org/10.1128/AAC.48.4.1289-1294.2004.
-
38.
Weinstein ZB, Zaman MH. Evolution of rifampin resistance in escherichia coli and mycobacterium smegmatis due to substandard drugs. Antimicrob Agents Chemother. 2019;63(1). [PubMed ID: 30397062]. [PubMed Central ID: PMC6325213]. https://doi.org/10.1128/AAC.01243-18.