1. Background
Early-onset neonatal sepsis (ENOS) is one of the most common causes of mortality in neonates (1). The bacteria causing ENOS are generally transferred from the mother to the infant before or during labor (1). By definition, ENOS is conceived to occur within the first 72 h of life in infants admitted to the neonatal intensive care unit or within the first seven days of life in term neonates (2-4). The incidence of ENOS in neonates with very low birth weight is estimated to be 15 - 24 per 1000 (5). Group B streptococci (GBS) and Escherichia coli are the leading causes of early-onset sepsis. Research indicates that the rectum or genital tract of 10 - 40% of pregnant women is colonized by GBS. There is a 30 - 70% chance of GBS transfer from colonized mothers to infants before or during childbirth, with 1 to 3% of the cases leading to severe disease (6).
Antibiotic administration during delivery conceivably reduces the incidence of GBS infections in term and preterm neonates (7). With a decreased incidence of GBS as a major contributor to the onset of sepsis (8), the role of Gram-negative bacteria, such as E. coli in particular, has increased (1, 9, 10). In a study by Tameliene et al., genitalia colonization with E. coli was reported in 7 - 13% of pregnant women. Escherichia coli was detected in the blood culture of 21% of their stillbirth fetuses (11). In recent years, in addition to E. coli, coagulase-negative staphylococci (CoNS) have played a major role in the development of EONS, especially in very low birth weight infants (5). By estimation, 9.3 and 2.5% of infants are colonized at birth with S. aureus and methicillin-resistant S. aureus (MRSA), respectively (12). Coagulase-negative staphylococci are reported in the oral cavity of 88.89% of newborns at 24 - 53 hours of birth (13). Some centers consider CoNS as real infections (1, 10, 14); however, others regard them as contamination (15-17).
2. Objectives
This study aimed to determine the prevalence rate of nasopharyngeal colonization of newly born infants with common bacterial agents causing ENOS, especially CoNS, and their relationship with blood culture outcomes in neonates admitted to a neonatal intensive care unit.
3. Methods
3.1. Patients
This cross-sectional, prospective study was performed in the neonatal intensive care unit of Imam Khomeini hospital in Ahvaz, the capital of Khuzestan province in the southwest of Iran with 34 beds, from December 2018 to April 2019. All neonates transferred to the neonatal intensive care unit for any reason were included in the study if they met the inclusion criteria. Neonates born with vaginal delivery, neonates born with cesarean sections, with mother’s fever (axillary temperature) above 38°C, membrane rupture ≥ 18h, preterm rupture of membrane, leukocytosis, and odorous vaginal discharge were included. Newborns requiring resuscitation at birth, neonates with a severe anomaly at birth, and those with a hospitalization duration of longer than 60 min were excluded.
3.2. Sampling and Culture Methods
Posterior pharynx secretions were swabbed, and the specimens were preserved in transport media. They were sent to the Microbiology Laboratory of Ahvaz University of Medical Sciences. Then, the specimens were inoculated on blood agar and MacConkey agar (Merck, Germany) and incubated at 37°C for 18 - 24 h (18). For blood culture, 1 ml of blood was taken from each neonate and inoculated into a blood culture bottle containing 10 mL of Trypticase Soy Broth (TSB) (Merck, Germany) and incubated at 37°C for 48 - 72 h (18).
3.3. Bacterial Identification
The grown colonies were examined by Gram staining, and then a culture medium was prepared by inoculating them in blood agar and MacConkey agar (Merck, Germany). The grown bacteria were identified by Gram staining, the type of hemolysis, and biochemical standard tests such as catalase, coagulase, DNase, bacitracin, and novobiocin sensitivity, Mannitol fermentation test, CAMP test, TSI, malonate, citrate, urea, SIM test, and gas production (18). The antibiotic sensitivity test was performed by the disk diffusion method (Kirby-Bauer) using Mueller-Hinton agar (Merck, Germany) according to the guidelines of the Clinical and Laboratory Standards Institute (CLSI) (19). All of these plates were incubated at 37°C for 18 - 24 h, and the clear zones of bacterial growth inhibition were measured and evaluated according to the CLSI guidelines (19).
3.4. Statistical Analysis
The quantitative variables were described as means, medians, and standard deviations. Frequency and percentage were employed to describe the qualitative variables. The normality of data distribution was assessed using the Q-Q and Kolmogorov-Smirnov tests. In cases where the variable conversion method was not possible, nonparametric tests were used. The Independent t-test, Mann-Whitney test, chi-square test, and Fisher’s exact test were used for data analysis. The data were analyzed using SPSS software, version 22, and the significance level was set at P ≤ 0.05.
4. Results
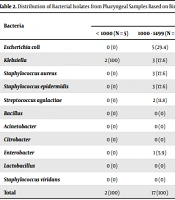
Of the 114 studied neonates, 59 (51.75%) were males, and 55 (48.24%) were females. The mean gestational age and birth body weight were 33.39 ± 3.48 weeks and 2,085.34 ± 692.24 g, respectively. The mothers’ mean age was 28.17 ± 6.13 years, and the mean number of pregnancies per woman was 2.68 ± 1.66. Moreover, 25 (21.9%) mothers had a history of abortion, and two (1.8%) had a history of previous child death. Over half (57.9%) of the deliveries were through the cesarean section. The pharyngeal culture was positive in 83 (72.8%) newborns (Table 1). Staphylococcus epidermidis was the most common bacterium isolated from pharyngeal samples in all weight groups (Table 2). Escherichia coli was resistant to ampicillin in 100% of the cases and gentamicin, cefotaxime, and ceftazidime in 50% of the cases (Table 3). Thirteen newborns died. Neonates’ pharyngeal samples were positive in 84.6% of the deceased infants and 71.28% of the surviving neonates (P = 0.882) (Table 4).
| Bacteria | No. (%) |
|---|---|
| Staphylococcus epidermidis | 32 (38.6) |
| Klebsiella | 12 (14.5) |
| Escherichia coli | 10 (12) |
| Staphylococcus aureus | 9 (10.8) |
| Streptococcus agalactiae | 6 (7.2) |
| Enterobacter | 4 (4.8) |
| Bacillus | 3 (3.6) |
| Citrobacter | 2 (2.4) |
| Lactobacillus | 2 (2.4) |
| Streptococcus viridans | 2 (2.4) |
| Acinetobacter | 1 (1.2) |
| Bacteria | Weight (%) | Total (%) | |||
|---|---|---|---|---|---|
| < 1000 (N = 5) | 1000 - 1499 (N = 31) | 1500 - 2499 (N = 54) | ≥ 2500 (N = 24) | ||
| Escherichia coli | 0 (0) | 5 (29.4) | 2 (4.9) | 3 (13) | 10 (12) |
| Klebsiella | 2 (100) | 3 (17.6) | 6 (14.6) | 1 (4.3) | 12 (14.5) |
| Staphylococcus aureus | 0 (0) | 3 (17.6) | 5 (12.2) | 1 (4.3) | 9 (10.8) |
| Staphylococcus epidermidis | 0 (0) | 3 (17.6) | 18 (43.9) | 11 (47.8) | 32 (38.6) |
| Streptococcus agalactiae | 0 (0) | 2 (11.8) | 2 (4.9) | 2 (8.7) | 6 (7.2) |
| Bacillus | 0 (0) | 0 (0) | 2 (4.9) | 1 (4.3) | 3 (3.6) |
| Acinetobacter | 0 (0) | 0 (0) | 1 (2.4) | 0 (0) | 1 (1.2) |
| Citrobacter | 0 (0) | 0 (0) | 1 (2.4) | 1 (4.3) | 2 (2.4) |
| Enterobacter | 0 (0) | 1 (5.9) | 2 (4.9) | 1 (4.3) | 4 (4.8) |
| Lactobacillus | 0 (0) | 0 (0) | 1 (2.4) | 1 (4.3) | 2 (2.4) |
| Staphylococcus viridans | 0 (0) | 0 (0) | 1 (2.4) | 1 (4.3) | 2 (2.4) |
| Total | 2 (100) | 17 (100) | 41 (100) | 23 (100) | 83 (100) |
| Antibiotics | Escherichia coli, % | Klebsiella, % | Staphylococcus aureus, % | Staphylococcus epidermidis, % | Streptococcus agalactiae, % |
|---|---|---|---|---|---|
| Ampicillin | 100 | 66.7 | 33.3 | 37.5 | 16.7 |
| Vancomycin a | - | - | 0 | 0 | 0 |
| Oxacillin | - | - | 11.1 | 15.6 | 0 |
| Imipenem | 20 | 25 | 22.2 | 18.8 | 0 |
| Meropenem | 10 | 25 | 6.3 | 6.3 | 0 |
| Amikacin | 40 | 33.3 | 33.3 | 18.8 | 0 |
| Gentamicin | 50 | 75 | 55.6 | 34.4 | 0 |
| Co-trimoxazole | 40 | 58.3 | 33.3 | 37.5 | 33.3 |
| Cefotaxime | 50 | 83.3 | 55.6 | 31.3 | 0 |
| Ceftazidime | 50 | 66.7 | 40.6 | 40.6 | 50 |
| Colistin b | 0 | 0 | - | - | - |
a Only Gram-positive bacteria were assessed.
b Only Gram-negative bacteria were assessed.
| Bacteria | Cases, No. (%) |
|---|---|
| Negative | 2 (15.38) |
| Escherichia coli | 3 (23.07) |
| Staphylococcus aureus | 3 (23.07) |
| Staphylococcus epidermidis | 3 (23.07) |
| Streptococcus agalactiae | 2 (15.3) |
| Total | 13 (100) |
Twelve neonates had positive blood cultures: (1) S. epidermidis in eight (66.7%); (2) E. coli in two (16.6%); and (3) Acinetobacter in two (16.6%) cases. Cultured S. epidermidis was resistant to ampicillin in 100% and sensitive to vancomycin in 100%. Besides, E. coli was resistant to ampicillin in 30% and sensitive to meropenem and amikacin in 100%. Also, Acinetobacter was resistant to ampicillin in 100%, meropenem in 50%, and amikacin in 100% of the cases. Simultaneous positive cultures from both blood and pharyngeal specimens were reported in seven (6.14%) cases. Also, the bacterial isolates in three (37.5%) of the blood and pharyngeal cultures were similar. Out of 114 cases, only three (2.6%) had similar pharyngeal and blood cultures (Table 5).
| Blood Isolates | Pharyngeal Isolates | Cases, No. (%) |
|---|---|---|
| Staphylococcus epidermidis | S. epidermidis | 1 (8.33) |
| S. epidermidis | Klebsiella | 2 (16.66) |
| S. epidermidis | Citrobacter | 1 (8.33) |
| S. epidermidis | Negative | 4 (33.33) |
| Escherichia coli | E. coli | 2 (16.66) |
| Acinetobacter | S. aureus | 1 (8.33) |
| Acinetobacter | Negative | 1 (8.33) |
| Total | 12 (100) |
Two of the 12 neonates with positive blood cultures died: (1) one was positive for E. coli; and (2) the other for Acinetobacter. The neonate with positive blood culture for E. coli also showed a positive pharyngeal culture for the bacterium. However, their strains were not determined. The pharyngeal culture was positive in 34 (64.2%) neonates with membrane rupture, 23 (79.3%) in infants with rupture < 18 h, and 26 (81.3%) in infants with rupture ≥ 18 h. The risks of positive pharyngeal culture in neonates with ruptures < 18 h and ≥ 18 h were 2.14 and 2.42 times, respectively. Also, the risk of positive pharyngeal culture was 1.13 higher in neonates with the membrane rupture ≥ 18 h compared to those with < 18 h (P = 0.151). Multiple logistic regression showed a 2.28 higher chance of positive pharyngeal culture in cases with membrane rupture than in those without membrane rupture, with the correlation being statistically significant (P = 0.053).
In terms of free oxygen intake, the chance of receiving oxygen in the negative pharyngeal culture group was 1.7 times higher than that in the positive pharyngeal culture group, although the difference was not statistically significant (P = 0.302). The use of nasal NCPAP was 1.86 times more in the negative pharyngeal culture group than in the positive pharyngeal culture group, although the difference was not statistically significant (P = 0.157). The chance of using mechanical ventilation was 1.74 times higher in the positive pharyngeal culture group than in the negative pharyngeal culture group, but the difference was not statistically significant (P = 0.426) (Table 6).
| Variables | β | S.E. | Wald | OR | P-Value |
|---|---|---|---|---|---|
| Rupture of membranes | 0.696 | 0.387 | 3.232 | 2.28 | 0.053 |
| Oxygen Therapy | -0.162 | 0.223 | 0.528 | 1.7 | 0.302 |
| Nasal CPAP | -0.416 | 0.232 | 3.214 | 1.8 | 0.157 |
| Mechanical Ventilation | 0.210 | 0.282 | 0.554 | 1.74 | 0.426 |
Other variables such as sex, weight, gestational age, type of delivery, mother’s age, number of pregnancies, duration of hospitalization, and white blood cell count of patients were not significantly different between the positive and negative pharyngeal culture groups.
5. Discussion
Our study showed no significant association between pharyngeal and blood bacterial culture (P = 0.73). Several large neonatal centers in the US used to culture specimens taken from different neonatal surfaces at birth, on the assumption that neonatal colonization is a precursor of neonatal sepsis and that positive bacterial culture is a cause of invasive infection (20-22). Numerous studies have shown that bacterial cultures from the axilla, groin, ear canal, and stomach secretions have poor predictive values for the diagnosis of early-onset sepsis and are therefore of limited value (23-25). However, some studies have demonstrated that nasopharyngeal culture has the highest positive predictive value among cultures of other body surfaces (25, 26). One study showed that the probability of clinical sepsis in colonized infants was significantly higher than that in non-colonized infants (27).
In our study, out of 114 neonates admitted to the neonatal intensive care unit, six (7.2%) were colonized with GBS. However, no cases of positive GBS blood cultures were found in the infants. While the colonization of pregnant women with GBS is of a substantial prevalence in Iran (28), neonatal sepsis with GBS is uncommon, and Gram-negative bacteria stand as the leading cause of neonatal infection (29). The use of CDC guidelines since 1996 concerning the use of antibiotic prophylaxis in mothers colonized with GBS has led to a remarkable reduction in neonatal infections with these bacteria (30). In this study, S. epidermidis was isolated from 38.6% of pharyngeal swabs and 66.7% of blood samples of neonates. Out of these isolates, 37.5% were resistant to ampicillin, and 100% were sensitive to vancomycin. The prevalence of CoNS in the development of early-onset sepsis ranges from 22.5 to 72.7% (27-29). This may be partly due to contamination during sampling for culture or due to the widespread presence of this group of bacteria on human skin. Although S. epidermidis is a bacterium with low pathogenicity in individuals with normal immune systems, it can cause invasive infections in infants, especially those with low birth weight.
In our study, 10 neonates were colonized with E. coli, and this bacterium was also isolated from blood samples of two (20%) cases of them. In Tameliene et al. study, the transmission rate from colonized mothers with E. coli to their neonates was 21.3%. In their study, the blood cultures of neonates were not assessed. Tameliene et al. (2012) showed also that the transmission rate from colonized mothers with E. coli to their neonates was 21.3%. In this study, the blood cultures of neonates were not assessed (11). The antibiotic susceptibility test in our study showed that 100 E. coli isolates from neonatal pharynges were resistant to ampicillin, and about 50% of the cases were resistant to gentamicin, cefotaxime, and ceftriaxone. Tameliene et al. (2012) in a similar study showed that 3.77% of E. coli isolates were resistant to ampicillin while 100% of them were sensitive to gentamicin and cefotaxime (11).
In some studies, the E. coli resistance rate to gentamicin in neonatal sepsis was 3%, while the resistance rate to ampicillin was between 30 and 85% (31-33). The studies in other parts of Iran also showed that the resistance rate was 100% to ampicillin and 0 to 33% to gentamicin (31-35). A study (2011) at our center reported that 100% of E. coli isolates were resistant to ampicillin and 82% to gentamicin (31). The indiscriminate use of ampicillin and gentamicin probably underlies the entry of resistant E. coli into the country. Although in our study, E. coli was isolated from two cases of blood and pharyngeal cultures, their serotypes were not determined. Therefore, it could not be established if neonatal sepsis originated from pharyngeal colonizing E. coli.
The rate of positive pharyngeal cultures was higher in neonates with membrane rupture of longer than 18 h than in newborns without membrane rupture or neonates with membrane rupture less than 18 h. Although the difference was not statistically significant, it showed that a longer membrane rupture increases the chances of developing colonization. There was no significant difference in the mortality rate between the colonized and non-colonized infants in this study. There was no significant difference between the colonized and non-colonized groups in terms of the clinical course and the need for mechanical ventilation. The low number of subjects to assess the incidence rate of sepsis associated with nasopharyngeal colonization with E. coli and the non-assessment of their serotypes were among the limitations of this study.
5.1. Conclusions
The results of this study showed that many babies are colonized at birth, but their colonization is not significantly associated with sepsis, mortality, blood cultures, or the need for medical care. Empirically, the treatment of neonates with suspected sepsis initiates with gentamicin and ampicillin. The high resistance of E. coli, which was the most common bacterium after S. epidermidis and Klebsiella, to ampicillin and a degree to gentamicin is a serious threat in the treatment of neonates with sepsis in our center. In addition, S. epidermidis was the most common isolate from blood and pharynx specimens that showed high resistance to ampicillin and gentamicin. These results suggest that the choice of antibiotics for the treatment of infants with suspected early-onset sepsis may need to be reconsidered.