Abstract
Background:
Pseudomonas aeruginosa is a nosocomial pathogen, acquiring resistance to a wide range of antibiotics. The MexAB-OprM pump can lead to resistance in this organism. Thus, the study was conducted to determine the effect of chitosan and phenylalanine arginyl ß-naphthylamide (PaβN) on the expression of MexAB in isolated ciprofloxacin resistant P. aeruginosa.Objectives:
This study investigated the effect of an antibiotic combination on the MexABP. aeruginosa expression.Methods:
A total of 30 ciprofloxacin-resistant isolates of P. aeruginosa were collected in this project. Then, chitosan nanoparticles were prepared using the ionic gelation method. Minimum inhibitory concentration (MIC) values were determined for ciprofloxacin, ciprofloxacin + PAßN, chitosan + ciprofloxacin, and chitosan + ciprofloxacin + PAßN using the micro-dilution method. Moreover, the expression level of MexAB genes was measured using real-time polymerase chain reaction.Results:
In total, 76.7% of the isolates were identified as multidrug resistant. A significant decrease in the MIC value was observed in groups treated with PAβN compared to those without PAβN. Moreover, the MIC value was significantly lower in the ciprofloxacin chitosan group than in groups without ciprofloxacin. Decreased MexA and MexB mRNA levels were observed in all antibiotic-treated strains compared to the ciprofloxacin-treated group.Conclusions:
There is a significant relationship between the increased MexAB expression and resistance to ciprofloxacin (P-value < 0.05). One of the therapeutic concerns is multidrug resistant bacteria, which needs to be addressed by finding new and more effective antibiotics.Keywords
Pseudomonas aeruginosa Phenyl-Arginine-Beta-Naphthylamide Chitosan Multidrug Efflux Pump Genes
1. Background
Pseudomonas aeruginosa is a vital Gram-negative bacterium that causes nosocomial infection. Hence, researchers nowadays consider it a challenging microorganism due to its durability on medical devices, such as catheters, and its ability to make biofilm on such surfaces (1). For this reason, the World Health Organization specified the organism as an important bacterium with different capabilities that needs a new drug for omitting the organism from clinical samples (2). Ciprofloxacin is a member of the fluoroquinolone class of antibiotics. It has been successfully used to treat many bacterial infections and is an effective antibiotic in treating P. aeruginosa infections (3). Efflux systems can cause antibiotic resistance to multiple antimicrobial agents. An antibiotic flow induced by Mex efflux proteins is the primary mechanism of antibiotic resistance in P. aeruginosa (4).
The presence of such efflux pumps in P. aeruginosa is the main reason for its resistance, which is controled by regulator genes (2). Due to the presence of the xenobiotic extrusion transporter of Mex-AB-OprM in this organism, it shows broad resistance to diverse types of antibiotics. Indeed, MexB, MexA, and OprM, coded by the MexAB-oprM operon, are responsible for this resistance (5). Resistance nodulation division (RND) pumps are involved in causing bacterial infections, and efflux pump inhibitors (EPIs) can be used as antimicrobial agents. In fact, the in vivo virulence of P. aeruginosa PAO1, either clinical or laboratory strains, could decrease by EPI Phe-Arg-βnaphthylamide (PAβN). PAβN is an active inhibitor of P. aeruginosa RND efflux pumps. This can help use fluoroquinolone in the over-expressed MexAB-OprM P. aeruginosa strain (6). The worldwide overuse of antibiotics and the presence of antibiotic-resistant microorganisms have encouraged scientists to design and develop new types of antibiotics. Nowadays, a combination of micro and nano delivery materials with antibiotics is used as an effective strategy for antibiotic-resistant microorganisms. Indeed, these materials protect antibiotics from destruction by enzymes and increase the drug's effectiveness (7).
Chitosan nanoparticles can go over biological barriers to keep macromolecules from degradation in the biological environment. Chitosan can also deliver drugs or macromolecules by controlled release to the target site. Also, the small size of chitosan makes it effective in interfacial action and reaction with the cell membrane because small particles are taken up by the cell via endocytosis. Many studies reported the ability of chitosan nanoparticles to enhance the bio access of drugs, changing their pharmacokinetics, and protect encapsulated drugs (8).
2. Objectives
This study was conducted to determine the effect of chitosan and PAβN on the MexA and MexB expression in clinical isolates of ciprofloxacin-resistant P. aeruginosa.
3. Methods
3.1. Isolation and Identification
A total of 151 P. aeruginosa isolates were detected from clinical specimens, such as trachea, urine, wound, CSF, and blood, from the Shahid Mohammadi Teaching Hospital, Bandar Abbas City, south of Iran. Bacterial isolates were identified using Gram staining, growth on the MacConkey agar, and biochemical tests, such as citrate utilization, oxidation-fermentation, and the oxidase test. Then, ciprofloxacin-resistant isolates were screened by 5µg ciprofloxacin disk using the disk diffusion method according to the CLSI guideline (9).
3.2. Antimicrobial Susceptibility Test
All the 30 ciprofloxacin-resistant isolates were subjected to the antimicrobial susceptibility test using disk diffusion assay. To this end, the Mueller-Hinton agar was used based on the Clinical and Laboratory Standards Institute (CLSI) references with 100/10 µg piperacillin-tazobactam, 30 µg ceftazidime, 10 µg imipenem, 10 µg meropenem, 30 µg aztreonam, 30 µg amikacin, and 10 µg tobramycin (Padtan Teb, Iran) (10).
3.3. Preparation of Chitosan Ciprofloxacin-Loaded Nanoparticles
In this project, the ionic gelation method was used to prepare chitosan nanoparticles. For this purpose, ciprofloxacin was dissolved in acetic acid (in 1% v/v) and supplemented with the chitosan solution. The nanoparticles were prepared by adding 0.25% sodium tripolyphosphate aqueous solution to the chitosan aqueous solution dropwise under 6000 rpm stirring speed at room temperature, until faint turbidity, analysis experiments were performed (11).
3.4. Minimum Inhibitory Concentration
Minimum inhibitory concentration (MIC) values for ciprofloxacin, ciprofloxacin + PAβN, chitosan + ciprofloxacin, and chitosan + ciprofloxacin + PAβN were measured using the microdilution method in 96 well plates. The concentration ranged between 0.25 - 256 µg/mL for ciprofloxacin (Fluka Analytical, Busch, Switzerland) and was 40 mg/L for PAβN (Sigma-Aldrich) after adding 100 µL of bacterial suspension with a population of 1.5 × 105 CFU/mL, incubated at 37°C for 24 h. MIC was determined as the lowest concentration of each compound, which inhibited bacterial growth (12).
Resistance due to the efflux pump with 2log2 dilutions higher than the wild strain type was considered as the MIC of the given strain in phenotypic analysis (13).
3.5. Molecular Analysis
3.5.1. DNA Extraction
In this study, the modified TE buffer boiling method was used to extract DNA. Thus, for each sample, five colonies of fresh bacteria were suspended in 200 µL of TE buffer [10 mM/L Tris-HCl, 1 mM/L EDTA (pH 7.5)]. The samples were centrifuged three times at 8000 rpm for 4 min at 4°C. The pellet was mixed in 200 µL TE buffer using a vortex mixer, and continued to boil in a heating block (Boeco, Germany) at 100°C for 1 min. Three freeze cycles were performed alternating between -70°C for 3 min and 100°C for 2 min. The tubes were centrifuged at 10000 rpm for 5 min (1). For quality control purity, the concentration was measured at the A260/A280 ratio with a Thermo Scientific NanoDropTM 1000 spectrophotometer (Thermo Scientific, Germany) using 1 µL of each sample (14). Finally, the supernatants were stored in a sterile tube at -20°C.
3.5.2. Polymerase Chain Reaction Amplification
Polymerase chain reaction (PCR) was carried out to detect MexA and MexB genes. For this purpose, 2 µL of DNA, 1× PCR buffer, 2 mM MgCl2, 200 µM of dNTP, 0.2 µM of each primer (Gen Fan Avaran, Iran) (Table 1), and 1 U Taq polymerase (SinaClon, Iran) were used to create the final reaction of 25 µL. The PCR condition was performed according to the following protocol: initial denaturation at 95°C for 5 min, 35 cycles of denaturation at 95°C for 30 s, annealing of MexA at 62°C and MexB at 59°C for 35 s, extension at 72°C for 40 s, and a final extension step at 72°C for 5 min. Afterward, each PCR product was mixed with 5 × Gel Red (Biotium, USA) and separated using electrophoresis on 1.2% agarose gel in TBE buffer (7.5 g EDTA, 108 g Tris and 55 g boric acid). After electrophoresis, a UV transilluminator (Proteinsimple USA) was used for visualization of the gel. In this project, the PCR outcome was considered as a positive control for the next steps, and the sequences were submitted to GenBank.
3.6. Gene Expression Analysis by Real-Time Reverse Transcription
The bacteria were cultured in a nutrient broth medium, with a sub-MIC concentration of ciprofloxacin (1 µg/mL) for each group of drugs (ciprofloxacin, chitosan + ciprofloxacin, ciprofloxacin + PAβN, and chitosan + ciprofloxacin + PAβN). The tubes were incubated at 37°C for 8 h. Total RNA was extracted from bacterial cells using an RNA extraction kit (Yekta Tajhiz Company, Iran) and then transcribed to cDNA with a cDNA synthesis kit (Yekta Tajhiz Company, Iran). The relative MexA and MexB expression was determined by qRT-PCR in triplicate using the Ampliqon SYBR Green kit and normalized to rpsL with forward and reverse primers shown in Table 1.
| Product Size | Reverse | Forward | Target |
|---|---|---|---|
| 316 bp | GGAGACCTTCGCCGCGTTGTCGC | CGACCAGGCCGTGAGCAAGCAGC | MexA |
| 273 bp | ACCTGGGAACCGTCGGGATTGA | CAAGGGCGTCGGTGACTTCCAG | MexB |
| 194 bp | CAAAACTGCCCGCAACGTC | TCCTTTACACGACCGCCAC | rpsL |
The primary protocol was as follows: initial denaturation at 94°C for 10 min, 40 cycles of amplification, denaturation at 94°C for 60 s, annealing at 61°C for 60 s, and extension at 72°C for 10 s (MexA, MexB, rpsL) (15). The relative expression level of efflux pump genes was calculated by the level of relative quantification in each efflux pump gene divided by that of rpsL (16).
3.7. Statistical Analyses
Statistical analyses were performed using SPSS (Version.22 IBM). One-way ANOVA (Tukey) was used to determine the statistical significance of the data, and P-value < 0.05 was considered statistically significant.
4. Results
4.1. Bacterial Strains
During the study period, a total of 151 P. aeruginosa isolates were recovered, of which 30 isolates were resistant to ciprofloxacin. The frequency of bacteria from each sample was as follows: (1) urine (43.3%), (2) tracheal aspirates (20%), (3) wound (16.7%), (4) blood (10%), and (5) secretions (10%).
4.2. Antimicrobial Susceptibility Test
As shown in Table 2, a high resistance rate was observed for piperacillin-tazobactam and meropenem (70%). Also, a low resistance rate was observed for tobramycin (40%). Furthermore, eight isolates (26.66%) were resistant to all the antibiotics. Overall, 76.7% of the isolates were recognized as multidrug resistant (MDR) due to at least three different classes of antibiotics.
Antimicrobial Susceptibility Test Results by Disk Diffusion on Pseudomonas aeruginosa Isolates
| Disk | Resistant (%) | Intermediate (%) | Susceptible (%) |
|---|---|---|---|
| Piperacillin–tazobactam | 70 | - | 30 |
| Ceftazidime | 60 | 26.7 | 13.3 |
| İmipenem | 50 | 6.7 | 43.3 |
| Meropenem | 70 | 16.7 | 13.3 |
| Aztreonam | 46.7 | 20 | 33.3 |
| Amikacin | 46.7 | 10 | 43.3 |
| Tobramycin | 40 | - | 60 |
4.3. Chitosan Ciprofloxacin-Loaded Nanoparticles
Ciprofloxacin loading did not cause any change in the morphology or size of chitosan, and ciprofloxacin was successfully encapsulated in chitosan. The ciprofloxacin loading efficiency was 35.51%. The release of the drug from chitosan nanoparticles was 53% after 4 hours, and more than 80% of the drug was released in 24 hours.
4.4. Minimum Inhibitory Concentration of Ciprofloxacin
Of the 151 P. aeruginosa isolates, 30 (19.87%) showed in vitro resistance to ciprofloxacin. Three of the isolates (10%) exhibited the MIC of 128 µg/mL, and nine of the isolates (30%) indicated the MIC of 64 µg/mL. The median MIC of ciprofloxacin varied between 32 and 4 µg/mL. The MIC range is shown in Table 3.
MIC and MBC of Ciprofloxacin Tested Clinical Isolates of Pseudomonas aeruginosa
| Number of Isolates | MIC (µg/mL) | MBC (µg/mL) | MBC/MIC Ratio |
|---|---|---|---|
| 1 | 128 | 256 | 2 |
| 1 | 64 | 256 | 4 |
| 8 | 64 | 128 | 2 |
| 2 | 32 | 128 | 4 |
| 1 | 32 | 64 | 2 |
| 1 | 16 | 64 | 4 |
| 1 | 16 | 32 | 2 |
| 1 | 8 | 32 | 4 |
| 3 | 8 | 16 | 2 |
| 7 | 4 | 8 | 2 |
| 2 | 4 | 4 | 1 |
4.5. Inhibitory Effect of PAβN on MexAB Efflux System
The MIC values were detected in terms of existence and non-existence of 40 mg/L of the efflux inhibitor PAβN. As shown in Table 4, MIC decreased by adding the inhibitor, and the difference in MIC between ciprofloxacin alone and with the inhibitor decreased 2-fold or more in all the samples. The results showed a significant reduction (P-value < 0.05) from 2- to 32-fold after using 40 µg.mL-1 of PAβN among the ciprofloxacin-resistant isolates. Also, of the isolates, three (10%) showed a 32-fold reduction, four (13.33%) a 16-fold reduction, eight (26.66%) an 8-fold reduction, six (20%) a 4-fold reduction, and nine (30%) a 2-fold reduction in MIC. There were no major differences between the MIC values in 21 out of the 40 ciprofloxacin-resistant isolates before and after using PAβN. The pattern of activity by PAβN showed that MexAB-OprM was active in 21 (70%) strains when MIC was performed after adding PAβN.
MICs of Ciprofloxacin with and Without PAβN on Pseudomonas aeruginosa Isolates
| Number of Isolates | Ciprofloxacin (µg/mL) | Ciprofloxacin with PAβN (µg/mL) | Fold Reduction |
|---|---|---|---|
| 2 | 128 | 64 | 2 |
| 1 | 128 | 32 | 4 |
| 2 | 64 | 32 | 2 |
| 5 | 64 | 16 | 4 |
| 2 | 64 | 2 | 32 |
| 3 | 32 | 16 | 2 |
| 1 | 16 | 8 | 2 |
| 1 | 16 | ≤ 0.5 | 32 |
| 4 | 8 | ≤ 0.5 | 16 |
| 1 | 4 | 2 | 2 |
| 8 | 4 | ≤ 0.5 | 8 |
4.6. The Minimum Inhibitory Concentration of Chitosan Loaded Drugs
We investigated the antibacterial activities of ciprofloxacin-chitosan and chitosan-ciprofloxacin-PAβN using the micro broth dilution test. The MIC values were significantly lower in the ciprofloxacin-chitosan than in free ciprofloxacin (P-value < 0.05). All results are summarized in Table 5.
Minimum Inhibitory Concentration Reduction Level of Ciprofloxacin-Resistant Isolates
| Isolate Number | Ciprofloxacin (µg/ml) | Chitosan Ciprofloxacin (µg/mL) | Fold of Reduction | ciprofloxacin PAβN (µg/mL) | Chitosan CiprofloxacinPAβN (µg/mL) | Fold of Reduction |
|---|---|---|---|---|---|---|
| 29 | 64 | 4 | 16 | 16 | 8 | 2 |
| 30 | 128 | 4 | 32 | 32 | 8 | 4 |
| 60 | 64 | 4 | 16 | 16 | 8 | 2 |
| 65 | 64 | 8 | 16 | 16 | 16 | - |
| 71 | 64 | 8 | 16 | 16 | 4 | 4 |
| 96 | 64 | 4 | 16 | 16 | 4 | 4 |
4.7. Polymerase Chain Reaction Amplification
As reported in the nucleotide sequence database, the nucleotide sequences were acquired with the accession numbers MW205514 for MexA and MW245012 for MexB (Figure 1).
Agarose gel electrophoresis of amplicons of MexA (part a) and MexB (part b) genes of Pseudomonas aeruginosa. Lane L, 100bp DNA marker; 1, positive control MexA gene (316bp) and MexB gene (273bp); 2, negative control; 3 - 7, MexA and MexB genes of P. aeruginosa isolates; 8, isolates without MexA and MexB genes.
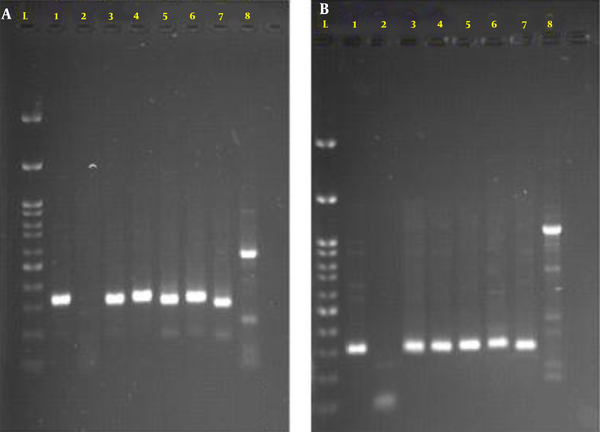
To determine the presence of MexA and MexB genes, the isolates with a 2-fold and more reduction in the MIC of ciprofloxacin with PAβN were subjected to PCR analysis. Six of the isolates showed a 2-fold reduction in the MIC of ciprofloxacin containing PAβN, and they were still in the resistance range (Table 5). These results explain the over-representation of MexA and MexB genes (83.33%) in our study.
4.8. MexA and MexB Expression Among Bacterial Strains
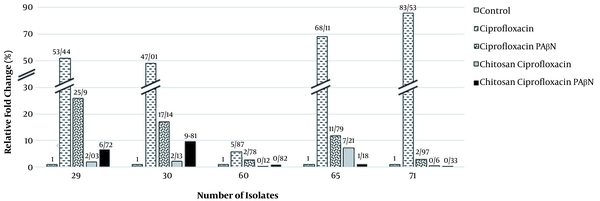
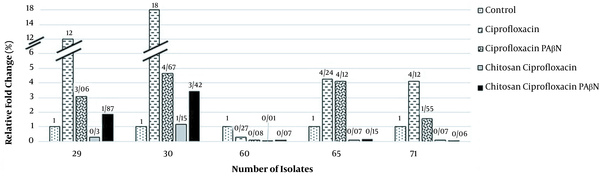
Our data indicated increased MexA and MexB mRNA levels in strains treated with ciprofloxacin. However, in strains exposed to ciprofloxacin with PAβN, the MexA and MexB transcription considerably decreased (Figures 2 and 3). Moreover, decreased MexA-MexB genes were observed in strains simultaneously exposed to chitosan-ciprofloxacin, but there were no substantial changes in strains treated with ciprofloxacin solely. Ciprofloxacin treatment greatly upregulated the MexA and MexB expression 51.59- and 7.72-fold on average, suggesting the activation of the MexA-MexB drug efflux pump system. On the contrary, ciprofloxacin-PAβN treatment slightly down-regulated the MexA and MexB expression. Thus, ciprofloxacin-PAβN treatment upregulated or suppressed the MexA-MexB system, which was activated by ciprofloxacin. Moreover, MexA and MexB genes were down-regulated 12.11- and 2.69-fold, respectively, with ciprofloxacin-PAβN treatment, compared to the control or ciprofloxacin-treated group.
Expression levels of MexA gene in ciprofloxacin-resistant Pseudomonas aeruginosa
Expression levels of MexB gene in ciprofloxacin-resistant Pseudomonas aeruginosa
5. Discussion
A deep understanding of ciprofloxacin resistance mechanisms could pave the way to improve the existing therapeutic regimes or even lead to developing new regimes that are effective against P. aeruginosa. Antimicrobial resistance is a worldwide health threat that needs to be investigated to develop new therapeutic strategies. Ciprofloxacin is used to treat many opportunistic infections caused by P. aeruginosa (3). Following the disk diffusion method, the highest resistance rate of 70% was observed in meropenem and piperacillin/tazobactam, while the lowest resistance rate of 40% was seen in tobramycin. In a study by Goudarzi in Iran, the frequency of MDR strains was 74.1%, which is close to our results of 76.7% (10). In a study by Pokhare et al., of all isolates, 28.2% were resistant to tobramycin, 23.9% to piperacillin/tazobactam, and 34.7% to meropenem (17). The antibiotic resistance of planktonic P. aeruginosa is mainly dependent on MDR pumps. Several MDR efflux pumps belong to P. aeruginosa and are coded by the RND family (18). PAβN is the most effective inhibitor for P. aeruginosa RND efflux pumps, which is discovered in a screen for adjuvants of fluoroquinolone levofloxacin, in a P. aeruginosa strain over-expressing MexAB-OprM (6).
In our study, of the isolates, 16 were found ciprofloxacin-resistant in the absence of an inhibitor, and 14 showed a significant 2-fold or more reduction in MIC in the presence of an inhibitor. Contrary to our study, Abbasi et al. reported 41 ciprofloxacin-resistant isolates, among which six showed a 2-fold or greater reduction in MIC (19). Furthermore, in accordance with our research, Farhangi showed a 4- and 2-fold increased antimicrobial activity of chitosan-loaded ciprofloxacin compared to free ciprofloxacin (20). Furthermore, Sobhani and his coworker illustrated that the MIC of ciprofloxacin-loaded chitosan nanoparticles was 50% lower than that of ciprofloxacin alone, which agrees with our results (11). In our study, the frequency of MexA and MexB efflux pumps was 83.33%. Our results showed that combination antibiotics (ciprofloxacin-PAβN and ciprofloxacin-chitosan) increased the bactericidal activity but considerably reduced the expression of MexA and MexB efflux pumps. We investigated the correlation between four types of drugs (ciprofloxacin, ciprofloxacin + PAβN, chitosan + ciprofloxacin, and chitosan + ciprofloxacin + PAβN) on expressions of efflux pump system genes (MexAB).
There was a direct correlation between the increased MexAB expression and resistance to ciprofloxacin (P-value < 0.05). The study respectively showed a 4.25- and 2.86-fold reduction in the MexA and MexB expression in strains simultaneously exposed to ciprofloxacin and PAβN, as compared to the ciprofloxacin treatment group. Also, the MexA expression decreased in the ciprofloxacin-PAβN treated group compared to the ciprofloxacin group. On average, a decrease in the MexA expression was observed by about 1/2. However, there was a significant expression reduction by about 1/4 in MexB, with a 2-fold reduction in some cases. Chitosan has a significant effect on reducing gene expression. Samples treated with ciprofloxacin and chitosan had a significant decrease in the MexA and MexB expression compared to those treated with ciprofloxacin alone. The ciprofloxacin and chitosan group had a higher effect on reducing gene expression than the ciprofloxacin and PAβN groups.
The MexA and MexB expression was reduced 21.33- and 24.14-fold after exposure to chitosan and ciprofloxacin, respectively. The expression of genes exposed to ciprofloxacin + chitosan + PAβN decreased 3.21- and 2.42-fold compared to genes exposed to ciprofloxacin + PAβN, but increased 1.55- and 3.48-fold compared to genes exposed to ciprofloxacin + chitosan, respectively. The reason might be inhibitory competition or antagonism with chitosan. The MIC results and the gene expression of different drug groups were shared. The chitosan and ciprofloxacin group was the most effective in efflux pump reduction, followed by the chitosan, ciprofloxacin, and PAβN group and the ciprofloxacin and PAβN group.
Our results also indicated a lower MexA and MexB expression among strains treated with chitosan + ciprofloxacin. However, the expression of both genes increased among strains exposed to chitosan + ciprofloxacin + PAβN. This finding is inconsistent with the results of Abdolhosseini et al. because they didn't use inhibitor during their experiment (21). In this study, attenuation of the gene expression of efflux pumps in ciprofloxacin resistance P. aeruginosa led to the synergism of PAβN with ciprofloxacin. In addition, the analysis illustrated a synergistic bactericidal potential in antibiotics and also in chitosan repressing microorganism growth in combined type. The leading causes of microbial inhibition cell lysis are the interference of chitosan with bacterial metabolism by electrostatic accumulation at the cell surface of bacteria and RNA transcription blockage from DNA by the uptake of chitosan into DNA molecules.
5.1. Conclusions
In conclusion, the relationship between chitosan and ciprofloxacin is promising in confronting multidrug-resistant bacteria. However, more research is required on the toxicity, efficacy, and stability of the combination of antimicrobial agents in in-vitro and in-vivo conditions. According to the results of this study, antibiotic resistance due to efflux pump overexpression is of great concern. Therefore, infection control with careful management of drug administration and identification of resistant isolates is essential to prevent the spread of resistance.
References
-
1.
Bahador N, Shoja S, Faridi F, Dozandeh-Mobarrez B, Qeshmi FI, Javadpour S, et al. Molecular detection of virulence factors and biofilm formation in Pseudomonas aeruginosa obtained from different clinical specimens in Bandar Abbas. Iran J Microbiol. 2019;11(1):25-30. [PubMed ID: 30996828]. [PubMed Central ID: PMC6462266].
-
2.
Housseini BK, Phan G, Broutin I. Functional mechanism of the efflux pumps transcription regulators from pseudomonas aeruginosa based on 3D structures. Front Mol Biosci. 2018;5:57. [PubMed ID: 29971236]. [PubMed Central ID: PMC6018408]. https://doi.org/10.3389/fmolb.2018.00057.
-
3.
Rehman A, Patrick WM, Lamont IL. Mechanisms of ciprofloxacin resistance in Pseudomonas aeruginosa: New approaches to an old problem. J Med Microbiol. 2019;68(1):1-10. [PubMed ID: 30605076]. https://doi.org/10.1099/jmm.0.000873.
-
4.
Adwan G, Abu-Shanab B, Adwan K. Antibacterial activities of some plant extracts alone and in combination with different antimicrobials against multidrug–resistant Pseudomonas aeruginosa strains. Asian Pac J Trop Med. 2010;3(4):266-9. https://doi.org/10.1016/s1995-7645(10)60064-8.
-
5.
Maseda H, Sawada I, Saito K, Uchiyama H, Nakae T, Nomura N. Enhancement of the mexAB-oprM efflux pump expression by a quorum-sensing autoinducer and its cancellation by a regulator, MexT, of the mexEF-oprN efflux pump operon in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2004;48(4):1320-8. [PubMed ID: 15047536]. [PubMed Central ID: PMC375252]. https://doi.org/10.1128/AAC.48.4.1320-1328.2004.
-
6.
Rampioni G, Pillai CR, Longo F, Bondi R, Baldelli V, Messina M, et al. Effect of efflux pump inhibition on Pseudomonas aeruginosa transcriptome and virulence. Sci Rep. 2017;7(1):11392. [PubMed ID: 28900249]. [PubMed Central ID: PMC5596013]. https://doi.org/10.1038/s41598-017-11892-9.
-
7.
Van Giau V, An SSA, Hulme J. Recent advances in the treatment of pathogenic infections using antibiotics and nano-drug delivery vehicles. Drug Des Devel Ther. 2019;13:327-43. [PubMed ID: 30705582]. [PubMed Central ID: PMC6342214]. https://doi.org/10.2147/DDDT.S190577.
-
8.
Divya K, Jisha MS. Chitosan nanoparticles preparation and applications. Environ Chem Lett. 2017;16(1):101-12. https://doi.org/10.1007/s10311-017-0670-y.
-
9.
Patel JB. Performance standards for antimicrobial susceptibility testing. Wayne, USA: Clinical and Laboratory Standards Institute; 2017.
-
10.
Moazami Goudarzi S, Eftekhar F. Multidrug resistance and integron carriage in clinical isolates of Pseudomonas aeruginosa in Tehran, Iran. Turk J Med Sci. 2015;45(4):789-93. [PubMed ID: 26422847]. https://doi.org/10.3906/sag-1408-120.
-
11.
Sobhani Z, Mohammadi Samani S, Montaseri H, Khezri E. Nanoparticles of chitosan loaded ciprofloxacin: Fabrication and antimicrobial activity. Adv Pharm Bull. 2017;7(3):427-32. [PubMed ID: 29071225]. [PubMed Central ID: PMC5651064]. https://doi.org/10.15171/apb.2017.051.
-
12.
Fuste E, Lopez-Jimenez L, Segura C, Gainza E, Vinuesa T, Vinas M. Carbapenem-resistance mechanisms of multidrug-resistant Pseudomonas aeruginosa. J Med Microbiol. 2013;62(Pt 9):1317-25. [PubMed ID: 23722434]. https://doi.org/10.1099/jmm.0.058354-0.
-
13.
Mesaros N, Glupczynski Y, Avrain L, Caceres NE, Tulkens PM, Van Bambeke F. A combined phenotypic and genotypic method for the detection of Mex efflux pumps in Pseudomonas aeruginosa. J Antimicrob Chemother. 2007;59(3):378-86. [PubMed ID: 17289770]. https://doi.org/10.1093/jac/dkl504.
-
14.
Abdel-Latif A, Osman G. Comparison of three genomic DNA extraction methods to obtain high DNA quality from maize. Plant Methods. 2017;13:1. [PubMed ID: 28053646]. [PubMed Central ID: PMC5209869]. https://doi.org/10.1186/s13007-016-0152-4.
-
15.
Sadredinamin M, Goudarzi H, Hashemi A, Tarashi S, Erfanimanesh S, Shariati A, et al. [Investigation of MexAB-OprM efflux pump expression level in Pseudomonas aeruginosa strains isolated from patients hospitalized in Shahid Motahari Hospital in Tehran City using real-time PCR method (Iran)]. Qom Univ Med. 2017;11(10):88-97. Persian.
-
16.
Shigemura K, Osawa K, Kato A, Tokimatsu I, Arakawa S, Shirakawa T, et al. Association of overexpression of efflux pump genes with antibiotic resistance in Pseudomonas aeruginosa strains clinically isolated from urinary tract infection patients. J Antibiot (Tokyo). 2015;68(9):568-72. [PubMed ID: 25850341]. https://doi.org/10.1038/ja.2015.34.
-
17.
Pokharel K, Dawadi BR, Bhatt CP, Gupte S. Prevalence of Pseudomonas aeruginosa and its antibiotic sensitivity pattern. J Nepal Health Res Counc. 2019;17(1):109-13. [PubMed ID: 31110388]. https://doi.org/10.33314/jnhrc.1877.
-
18.
Soto SM. Role of efflux pumps in the antibiotic resistance of bacteria embedded in a biofilm. Virulence. 2013;4(3):223-9. [PubMed ID: 23380871]. [PubMed Central ID: PMC3711980]. https://doi.org/10.4161/viru.23724.
-
19.
Abbasi F, Yusefi S, Yavar SA. Minimum inhibitory concentration of ciprofloxacin against Pseudomonas aeruginosa in the presence of the efflux inhibitor phenylalanine-arginine beta-naphthylamide. Intern Med Med Investig J. 2018;3(4):23-7. https://doi.org/10.24200/imminv.v2i4.128.
-
20.
Farhangi M, Kobarfard F, Mahboubi A, Vatanara A, Mortazavi SA. Preparation of an optimized ciprofloxacin-loaded chitosan nanomicelle with enhanced antibacterial activity. Drug Dev Ind Pharm. 2018;44(8):1273-84. [PubMed ID: 29452500]. https://doi.org/10.1080/03639045.2018.1442847.
-
21.
Abdolhosseini M, Zamani H, Salehzadeh A. Synergistic antimicrobial potential of ciprofloxacin with silver nanoparticles conjugated to thiosemicarbazide against ciprofloxacin resistant Pseudomonas aeruginosa by attenuation of MexA-B efflux pump genes. Biologia. 2019;74(9):1191-6. https://doi.org/10.2478/s11756-019-00269-0.