1. Background
Pseudomonas aeruginosa is a unique Gram-negative opportunistic pathogen that causes a broad range of complicated-to-treat infections that resist antibiotic therapies (1, 2). Pseudomonas aeruginosa has the remarkable capability to develop resistance to a broad class of antibiotics and is associated with significant morbidity and mortality (3, 4). Multidrug-resistance (MDR) in P. aeruginosa is associated with the prevalence of different genes, especially those encoding class B carbapenemases [Metallo-β-lactamases (MBLs)] and extended-spectrum β-lactamases (ESBLs) (5, 6). Since MBL and ESBL-producing bacteria are often MDR, the treatment of infections caused by these bacteria is very challenging (7). Clinicians have to use polymyxins, such as colistin and polymyxin B, as a last-line antimicrobial agent. However, recently published studies have reported that resistance to polymyxins, especially colistin, is increasing due to the spread of mcr-1 and other mcr genes (8).
Pseudomonas aeruginosa can acquire resistance to carbapenems and β-lactams with the production of carbapenemases, including the Verona integron-encoded β-lactamase (VIM), imipenemase (IMP), and ESBLs such as the TEM, SHV, and CTX-M type enzymes (9-11). Carbapenemase and ESBL enzymes can hydrolyze broad-spectrum β lactam antibiotics, including imipenem, meropenem, cephalosporins, cefotaxime, ceftazidime, and monobactams (12, 13). Ambler class B enzymes, such as VIM and IMP, are the most frequent enzymes related to the carbapenemases-mediated resistance mechanism, and in the case of ESBLs, TEM, SHV, and CTX-M types are the vast majority of enzymes among Gram-negative bacilli (7, 14).
2. Objectives
Given the importance of increasing resistance mediated by carbapenemases and ESBLs, this study aimed to investigate the prevalence of the main carbapenemases (blaVIM and blaIPM) and ESBLs (blaTEM, blaSHV, and blaCTX) encoding genes in P. aeruginosa clinical isolates collected from three military hospitals in Tehran, Iran.
3. Methods
3.1. Bacterial Isolates
In this cross-sectional study, we collected 85 P. aeruginosa isolates from hospitalized military patients in different wards of three university hospitals [501 Hospital (Imam Reza), Khanevadeh Artesh Hospital, and Besat General Hospital] in Tehran, Iran, from 2019 to 2020 (Table 1). At the first step, all bacterial samples were cultured on common bacterial growth media, including blood agar, MacConkey agar, and Tryptic Soy Broth, and incubated at 37ºC for 18 to 24 hours. After incubation, the conventional biochemical and microbiologic tests, including Gram stain, catalase, and oxidase test, growth at 42ºC, growth on triple sugar iron agar and Kligler iron agar, pigment production on Mueller-Hinton agar (Merck Co., Germany), IMVIC (Indole, Methyl red, Voges proskauer, and Citrate) test, and motility test were used to identify the isolates (2, 15). We stored the isolates at -70ºC in Trypticase soy broth (TSB) containing 20% glycerol until molecular analysis.
| Genes | Primer Sequences | Product Size (bp) |
|---|---|---|
| blaIMP | F: 5’- GGAATAGAGTGGCTTAATTCTC-3’; R: 5’- CCAAACCACTACGTTATCT-3’ | 189 |
| blaVIM | F: 5’-ATGGTGTTTGGTCGCATATC-3’; R: 5’-TGGGCCATTCAGCCAGATC-3’ | 510 |
| blaTEM | F: 5’-ATGAGTATTCAACATTTCCG-3’; R: 5’-CTGACAGTTACCAATGCTTA-3’ | 867 |
| blaSHV | F: 5’-GATGAACGCTTTCCCATGATG -3’; R: 5’-CGCTGTTATCGCTCATGGTAA -3’ | 214 |
| blaCTX | F: 5’-TTTGCGATGTGCAGTACCAGTAA -3’; R: 5’-CGATATCGTTGGTGGTGCCATA -3’ | 590 |
| mcr-1 | F: 5’- CGGTCAGTCCGTTTGTTC-3’; R: 5’- CTTGGTCGGTCTGTAGGG-3’ | 309 |
3.2. Antimicrobial Susceptibility Test
We carried out antimicrobial susceptibility testing using the disk diffusion method on Mueller-Hinton agar (Merck) according to the guidelines of the Clinical and Laboratory Standards Institute (CLSI). Here, the susceptibility of the isolates to the 12 antimicrobial agents of imipenem (IMI; 10 μg), meropenem (MEM; 10 μg), cefepime (CPM; 30 μg), cefotaxime (CTX; 30 µg), ceftazidime (CAZ; 30 μg), piperacillin (PRL; 100 μg), piperacillin/tazobactam (PTZ; 100/10 μg), ticarcillin (TIC; 75 μg), ciprofloxacin (CIP; 5 μg), gentamicin (GM; 10 μg), amikacin (AK; 30 μg), and colistin sulfate (CO; 10 μg) (Mast, UK) was determined. We classified the isolates resistant to at least three different antimicrobial agents as MDR (15). We also used the agar dilution method to determine the minimum inhibitory concentrations (MICs) of gentamicin, imipenem, ciprofloxacin, piperacillin, and colistin (16). The interpretation of MIC results was carried out according to the CLSI breakpoints (imipenem and colistin: susceptible: ≤ 2 μg/mL, resistant: ≥ 8 μg/mL, gentamicin: susceptible: ≤ 4 μg/mL, resistant: ≥ 16 μg/mL, piperacillin: susceptible: ≤ 16 μg/mL, resistant: ≥ 128 μg/mL, ciprofloxacin: susceptible: ≤ 1 μg/mL, resistant: ≥ 4 μg/mL) (17). We defined MIC as the lowest concentration of an antimicrobial agent that inhibited the bacteria's growth compared with P. aeruginosa PAO1 as the positive control.
3.3. Molecular Detection of Genes
According to the manufacturer, we extracted all the collected isolates' whole genomic DNA using DNJia Plus Tissue and Bacteria Kit’s (ROJE Technologies Co, Iran: Cat. No.: DN983051) protocol. Specific primers (Table 1) for the blaIPM, blaVIM, blaSHV, blaTEM, blaCTX, and mcr-1 genes were used for molecular detection using polymerase chain reaction (PCR) methods. The primers and their thermal cycling protocols used in this study were described previously (18-21). We carried out DNA amplification on a thermal cycler (Eppendorf, Mastercycler Gradient; Eppendorf, Hamburg, Germany) under the following condition: (1) 10 - 20 ng of extracted sample DNA; (2) 1 µL of each primer (10 pmol); (3) 12.5 µL of Taq DNA Polymerase 2x Master Mix RED (Ampliqon Co, Denmark; Cat. No.: A180301); and (4) up to 25 µL distilled water. Polymerase chain reaction conditions were set based on a previously published study by Azimi et al. (20). We assessed all PCR products by electrophoresis on 1% agarose gel (SinaClon, Iran). Escherichia coli ATCC 25922 was used as a positive control for the PCR assay.
4. Results
4.1. Number and Distribution of Isolates
From 2019 to 2020, 85 P. aeruginosa clinical strains were collected from three university hospitals [501 Hospital (Imam Reza), Khanevadeh Artesh Hospital, and Besat General Hospital] in Tehran, Iran. The distribution of P. aeruginosa isolates in different hospital wards and among different clinical samples is shown in Table 2. The patients' age ranged from 1 to 89 years (56.5 ± 22), and approximately 56.5 and 43.5% of the P. aeruginosa isolates belonged to female and male samples, respectively. Our analyses revealed that most P. aeruginosa isolates were identified in the ICU (n = 34; 40%) and surgery ward (n = 16; 18.8%), respectively. Moreover, the results revealed that most P. aeruginosa isolates were isolated from wound and urine samples, respectively.
| Variables | No. | % |
|---|---|---|
| Sex | ||
| Females | 48 | 56.5 |
| Males | 37 | 43.5 |
| Ward | ||
| Intensive care unit (ICU) | 34 | 40 |
| Surgery | 16 | 18.8 |
| Orthopedic | 5 | 5.9 |
| Internal | 11 | 12.9 |
| Emergency | 14 | 16.5 |
| General | 5 | 5.9 |
| Origin | ||
| Wound | 28 | 32.9 |
| Urine | 23 | 27 |
| Tracheal aspirate | 14 | 16.5 |
| Blood | 9 | 10.6 |
| Sputum | 6 | 7 |
| Fluid | 3 | 3.6 |
| Stool | 2 | 2.4 |
| Total | 85 | 100 |
4.2. Antimicrobial Susceptibility Test
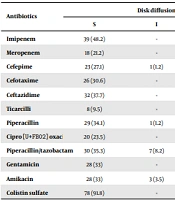
Table 3 summarizes the results of the disk diffusion test and the agar dilution method. Our results showed that all the isolates were resistant to two antibiotics, and 76 (89.4%) of isolates were MDR. The maximum and minimum resistance rates were against ticarcillin (n = 77; 90.5%) and colistin (n = 7; 8.2%), respectively. In carbapenem antibiotics, 76 (89.4%) isolates were resistant to at least imipenem or meropenem or both of them. Resistance to the cephalosporins group was higher (n = 79; 92.9%) than carbapenems. Data from MIC assay showed that among the five tested antibiotics, isolates had higher resistance to ciprofloxacin (n = 63; 74.1%). In general, our findings showed that from 34 P. aeruginosa isolated from the ICU, 21 (61.8%) P. aeruginosa isolates were resistant to meropenem and cefepime. Moreover, 12 (75%) P. aeruginosa isolates collected from the surgery ward were resistant to cefotaxime and ticarcillin. On the other hand, 21 (75%), 16 (69.5%), and 10 (71.4%) of P. aeruginosa isolates recovered from wound, urine, and tracheal aspirate samples were resistant to ticarcillin, respectively. Results showed that in comparison to other clinical samples, resistance rates to meropenem (82.1%), cefotaxime (67.8%), and amikacin (64.2%) among P. aeruginosa isolated from wound samples were high.
| Antibiotics | Disk Diffusion | Agar Dilution | ||||
|---|---|---|---|---|---|---|
| S | I | R | S | I | R | |
| Imipenem | 39 (48.2) | - | 44 (51.8) | 19 (29.5) | 7 (8.2) | 54 (62.3) |
| Meropenem | 18 (21.2) | - | 67 (78.8) | - | - | - |
| Cefepime | 23 (27.1) | 1 (1.2) | 61 (71.7) | - | - | - |
| Cefotaxime | 26 (30.6) | - | 59 (69.4) | - | - | - |
| Ceftazidime | 32 (37.7) | - | 53 (62.3) | - | - | - |
| Ticarcilli | 8 (9.5) | - | 77 (90.5) | - | - | - |
| Piperacillin | 29 (34.1) | 1 (1.2) | 55(64.7) | 22 (26.9) | 2 (2.4) | 61 (71.7) |
| Ciprofloxacin | 20 (23.5) | - | 65 (76.5) | 19 (22.4) | 3 (3.5) | 63 (74.1) |
| Piperacillin/tazobactam | 30 (35.3) | 7 (8.2) | 48 (56.5) | - | - | - |
| Gentamicin | 28 (33) | - | 57 (67) | 25 (29.4) | 1 (1.2) | 59 (69.4) |
| Amikacin | 28 (33) | 3 (3.5) | 54 (63.5) | - | - | - |
| Colistin sulfate | 78 (91.8) | - | 7 (8.2) | 74 (89.4) | 2 (2.4) | 7 (8.2) |
Abbreviations: S, sensitive; I, intermediate; R, resistant.
a Values are expressed as No. (%).
4.3. Distribution of MBL and ESBL-Producing Isolates
We carried out the PCR technique to detect blaIPM, blaVIM, blaSHV, blaTEM, and blaCTX, genes in all P. aeruginosa isolates. The frequency of this genes was as follow:
- MBL genes: blaVIM (n = 44; 51.8%), blaIPM (n = 20; 23.5%).
- ESBL genes: blaTEM (n = 41; 48.2%), blaSHV (n = 24; 28.2%), and blaCTX (n = 16; 18.8%). The frequency rates of MBL- and ESBL-producing isolates in this study were 56 (65.9%) and 60 (70.6%), respectively. Among all the isolates, 81 (95.3%) had at least one of the studied genes. Table 4 shows the distribution of these genes among all the P. aeruginosa strains collected in this study. Data analysis showed that the presence of the blaIMP and blaVIM genes was significantly associated with resistance to carbapenems (P < 0.002). Also, for ESBL genes, namely blaTEM, blaSHV, and blaCTX, this association was observed for cephalosporins (P < 0.007), including cefepime, cefotaxime, and ceftazidime. Due to the resistance to colistin observed in seven isolates, we also carried out PCR to detect the mcr-1 gene, but we did not find it in any isolates. Our analyses revealed that in comparison to other hospital wards, the frequency of blaVIM and blaTEM genes among the P. aeruginosa isolates collected from ICU and emergency wards was high. On the other hand, the frequency of these genes among P. aeruginosa strains isolated from the surgery, and internal wards was low. Moreover, the frequency of blaVIM and blaTEM genes among P. aeruginosa strains isolated from wound and urine samples was high.
| Resistance Genes | No. (%) |
|---|---|
| blaVIM | 44 (51.8) |
| blaIPM | 20 (23.5) |
| blaTEM | 41 (48.2) |
| blaSHV | 24 (28.2) |
| blaCTX | 16 (18.8) |
| Combination of genes a | |
| blaVIM | 11 (12.9) |
| blaIPM | 7 (8.2) |
| blaTEM | 6 (7) |
| blaSHV | 6 (7) |
| blaCTX | 3 (3.5) |
| blaVIM + blaIPM | 3 (3.5) |
| blaVIM + blaTEM | 13 (15.3) |
| blaVIM + blaSHV | 4 (4.7) |
| blaVIM + blaCTX | 3 (3.5) |
| blaIPM + blaTEM | 2 (2.4) |
| blaIPM + blaCTX | 1 (1.2) |
| blaTEM + blaSHV | 5 (5.9) |
| blaTEM + blaCTX | 2 (2.4) |
| blaSHV + blaCTX | 1 (1.2) |
| blaVIM + blaIPM + blaTEM | 2 (2.4) |
| blaIPM + blaTEM + blaCTX | 3 (3.5) |
| blaVIM + blaTEM + blaSHV | 5 (5.9) |
| blaVIM + blaIPM + blaSHV | 1 (1.2) |
| blaTEM + blaSHV+ blaCTX | 2 (2.4) |
| blaVIM + blaIPM + blaTEM + blaCTX | 1 (1.2) |
| None of studied genes | 4 (4.7) |
a Other combination of genes was not detected in this study.
5. Discussion
Metallo-β-lactamases- and ESBL-producing isolates of P. aeruginosa are the most frequent pathogen causing infections in hospitalized patients. Acquired resistance due to the widespread use of antibiotics, especially in clinical treatments, increases antimicrobial resistance in pathogenic bacteria with emerging MDR and extensively drug-resistant (XDR) and pan drug-resistant (PDR) strains (22, 23). Multidrug-resistant mechanisms of P. aeruginosa are mainly due to ß-lactamase production, efflux pumps, and outer membrane proteins changes (24). This study aimed to evaluate the prevalence of MBL, and ESBL genes in P. aeruginosa isolates recovered from patients in different wards of three military hospitals in Tehran, Iran. Data analysis showed that most of the isolates were MDR (89.4%), and the highest resistance was observed for ticarcillin (90.5%), followed by meropenem (78.8%) and ciprofloxacin (76.5%). As previous studies reported, the rate of resistance to antimicrobial agents varies in different areas of Iran, and antibiotics resistance has arisen significantly in P. aeruginosa over the past two decades (25).
Various studies have surveyed the prevalence of antibiotic resistance among P. aeruginosa strains. The results of a study performed by Azimi et al. revealed that P. aeruginosa had the lowest and highest resistance rates to levofloxacin and ticarcillin-clavulanic acid, respectively (15). Labaste et al. revealed that 20.5% of P. aeruginosa strains developed carbapenem resistance (26). Emaneini et al. reported that polymyxin B, piperacillin/tazobactam and meropenem were the most active antibiotics against P. aeruginosa isolates (27). In general, antimicrobial susceptibility and MIC test results are relatively similar to previous findings obtained by Farshadzadeh et al. from Iran, Goli et al. from Iran, and Adjei et al. from South Africa (28-30). However, we found more variation in resistance rates than in other studies (23, 31, 32). These differences in resistance rates are probably because of the accumulation of resistance to that area's antimicrobial agents.
Among all isolates, 95.3% had at least one of the studied genes. The most frequent genes in this study were blaVIM (n = 44; 51.8%) and blaTEM (n = 41; 48.2%), respectively. Also, our data showed that 65.9% and 70.6% of isolates were positive for MBL- and ESBL-producing genes. In a study by Salimi and Eftekhar on 32 carbapenem-resistant, MBL-producing P. aeruginosa isolates, 56.25 and 46.8% of the isolates were positive for blaIMP and blaVIM, respectively (33). However, in our results, the carriage for the blaVIM gene was more than two times higher than for blaIMP. Aghamiri et al. reported that the frequency of blaVIM (33%) was higher than blaIPM (9%), but their frequencies were lower than our results (34). In another study, Shahcheraghi et al. reported that 68% of MBL-positive strains of P. aeruginosa harbored blaVIM, but none was blaIMP-positive (35). In conflict with our findings, Radan et al. reported that 74.3% of MBL isolates were positive for the blaIMP gene (36).
In the case of ESBL genes, blaTEM, blaSHV, and blaCTX genes vary in different studies. Our results, like other studies in Iran, showed that the most prevalent ESBL gene was blaTEM. Pakbaten Toupkanlou et al. reported that among ESBL-positive strains of P. aeruginosa, the frequency of the blaTEM and blaSHV genes was 50% (22). Another report by Bokaeian et al. revealed that the blaTEM and blaSHV genes were harbored by 100 and 6.6% of ESBL-positive strains of P. aeruginosa, respectively (37). Bahrami et al. also reported that the prevalence rates of blaTEM, blaSHV, and blaCTX-M were 57.29, 23.08, and 23.95% among 96 isolates of P. aeruginosa (6).
5.1. Conclusion
Our results indicated that resistance to almost all available antibiotics used in clinical settings against P. aeruginosa is significantly increasing. Pseudomonas aeruginosa resistance to antimicrobial agents is due to several mechanisms, but MBL- and ESBL genes cause resistance to the most frequently used antibiotics against this pathogen. In sum, our results indicated that the frequency rates of MBL- and ESBL genes are high among P. aeruginosa isolates, which could be the reason for the increased resistance to antimicrobial agents.