1. Background
Immunosuppression is the most critical risk factor for the recurrence of vaginal infections (1); for example, women with cervical dysplasia may be susceptible to infection by certain pathogenic bacteria. Bacterial vaginosis (BV) occurs when lactobacilli, present in the vagina of healthy women, are replaced or overcome by Gram-negative, strict anaerobic, or facultative anaerobic bacteria present in small numbers (2). The genomic capacity of Escherichia coli allows the expression of numerous virulence factors responsible for vaginal infection (3). Some of the virulence factors produced by E. coli strains include toxins, siderophores, iron receptors, fimbriae, flagella, aphymic adhesins, and immune evasion mechanisms (4).
Antimicrobial action has been hampered by bacteria multidrug-resistant to certain antimicrobial families (5). Antimicrobial resistance genes are usually expressed in integrons. Class 1 genes are the most studied since they are found in Gram-negative strains of clinical origin, playing an essential role in bacterial resistance and in generating and expressing antibiotic resistance cassettes (6). A previous study of our group demonstraTed that the Class 1 integrons of E. coli of vaginal origin were reservoirs of antimicrobial resistance genes (7). However, there is still controversy about the role of this bacterium as a causal agent of this type of pathology since the presence of the main virulence genes associated with cytotoxicity and adhesion is not considered. Implications are discussed for identifying E. coli carrying resistance and virulence factors as a causative agent of BV in dysplastic women.
2. Objectives
This work aimed to provide additional experimental evidence by identifying virulence genes, phenotypes, and genotypes associated with Class 1 integrons that may carry genes conferring resistance to different families of antibiotics in commensal and virulent E. coli strains isolated from immunocompromised patients.
3. Methods
3.1. Study Population and Bacterial Isolation/Identification
This study included one hundred and five E. coli strains isolated from women with cervical dysplasia and monobacterial vaginal infection diagnosed with BV. The strains were collected from August 2019 to January 2020 at the Dysplasia Service and Research Division of the "Hospital Juarez de Mexico." Typical colonies were subjected to presumptive identification according to Cowan and Steel's criteria on LB agar (8). Strain identification was performed using BD PhoenixTM (Brea, California, USA) following the manufacturer's protocol. The resistance phenotype was confirmed according to the criteria specified by the Clinical and Laboratory Standards Institute (CLSI) as follows.
3.2. Resistance Phenotype Assay
The E. coli strains were subjected to antimicrobial resistance phenotyping. For this purpose, the disk diffusion test was performed according to the protocol established by CLSI (9) on the surface of Mueller-Hinton agar plates. Twelve antimicrobial agents were used to determine antimicrobial susceptibility: (1) 10 μg of ampicillin; (2) 100 μg of carbenicillin; (3) 30 μg of cephalothin; (4) 30 μg of cefotaxime; (5) 5 μg of ciprofloxacin; (6) 30 μg of chloramphenicol; (7) 300 μg of nitrofurantoin; (8) 30 μg of amikacin; (9) 10 μg of gentamicin; (10) 30 μg of netilmicin; (11) 10 μg of norfloxacin; and (12) 1 25/23.75 μg of trimethoprim/sulfamethoxazole (BD, Brea, California, USA). The antimicrobial susceptibility profile of the isolates was reported as susceptible or resistant.
3.3. DNA Extraction and Phylogenetic Assignment
The Wizard® Genomic DNA Purification Kit (Promega Corporation, Madison, USA) was used for total DNA extraction. Prior to phylogenetic assignment, the 16S rRNA gene was amplified to verify the quality and amplifiability of the DNA samples (7). Clermont et al. (10) proposed a dichotomous analysis based on the chuA, yjaA, and arpA genes and the anonymous fragment designated as TspE4.C2 proposing six main phylogenetic groups in E. coli strains of different origins. Multiplex PCR conditions were pre-denaturation for 4 min at 94°C, followed by up to 30 cycles of denaturation for 5 sec at 94°C, annealing for 20 sec at 59°C, and extension for 1 min at 72°C, with a final extension of 5 min at 72°C. PCR products were resolved on agarose gels and documented under UV light and ethidium bromide. The primers used for this purpose and the length of the PCR products are shown in Table 1.
| Molecular Target and Primers | Sequence (5’ to 3’) | Size (bp) | Anneling Temperature (oc) |
|---|---|---|---|
| 16S rRNA | 1492 | 57 | |
| 27-F | AGAGTTTGATCCTGGCTCAG | ||
| 1492-R | GGTTACCTTGTTACGACTT | ||
| Hemin uptake system | 279 | 59 | |
| chua-F | TGCCGCCAGTACCAAAGACA | ||
| chua-R | GACGAACCAACGGTTCAGGAT | ||
| Unknown | 211 | 59 | |
| yjaA-F | TGAAGTGTCAGGAGACGCTG | ||
| yjaA-R | ATGGAGAATGCGTTCCTCAAC | ||
| Anonymous DNA fragment | 152 | 59 | |
| TSPE4.C2-F | GAGTAATGTCGGGGCATTCA | ||
| TSPE4.C2-R | CGCGCCAACAAAGTATTACG | ||
| Ankyrin-like regulatory protein | 400 | 59 | |
| arpA-F | AACGCTATTCGCCAGCTTGC | ||
| arpA-R | TCTCCCCATACCGTACGCTA | ||
| 5´-Conserved segment “Integrase intI1” | 923 | 50 | |
| IntI1-F | GTTCGGTCAAGGTTCTG | ||
| IntI1-R | GCCAACTTTCAGCACATG | ||
| 3´-Conserved segment “quacΔE1/sul11” | 800 | 56 | |
| QacEΔ1-F | ATCGCAATAGTTGGCGAAGT | ||
| sul1-B | GCAAGGCGGAAACCCGCGCC | ||
| Variable region | Variable | 59 | |
| in-F | GGCATCCAAGCAGCAAGC | ||
| in-B | AAGCAGACTTGACCTGAT | ||
| Intergenic consensus | Variable | 50 to52 | |
| ERIC1R | ATGTAAGCTCCTGGGGATTCA | ||
| ERIC2 | AAGTAAGTGACTGGGGTGAGC | ||
| Serum resistance associated | 290 | 59 | |
| traT-F | GGTGTGGTGCGATGAGCACAG | ||
| traT-R | CACGGTTCAGCCATCCCTGAG | ||
| Adhesin | 508 | 59 | |
| fimH-F | TCGAGAACGGATAAGCCGTGG | ||
| fimH-R | GCAGTCACCTGCCCTCCGGTA | ||
| Aerobactin siderophore | 300 | 59 | |
| iutA-F | GGCTGGACATCATGGGAACTGG | ||
| iutA-R | CGTCGGGAACGGGTAGAATCG | ||
| Pyelonephritis associated pili | 200 | 59 | |
| papC-F | ATATCCTTTCTGCAGGGATGCAATA | ||
| papC-R | GTGGCAGTATGAGTAATGACCGTT | ||
| Cytotoxic necrotizing factor 1 | |||
| cnf1-F | AAGATGGAGTTTCCTATGCAGGAG | 498 | 63 |
| cnf1-R | CATTCAGAGTCCTGCCCTCATTATT | ||
| Alpha-hemolysin toxin | 1177 | 63 | |
| hlyA-F | AACAAGGATAAGCACTGTTCTGGC | ||
| hlyA-R | ACCATATAAGCGGTCATTCCCGTCA | ||
| Serin autotransporter toxins | 667 | 57 | |
| sat-F | TGCTGGCTCTGGAGGAAC | ||
| sat-R | TTGAACATTCAGAGTACCGGG | ||
| Serine protease autotransporter toxin | 409 | 58 | |
| pic-F | ACTGGATCTTAAGGCTCAGG | ||
| pic-R | TGGAATATCAGGGTGCCACT | ||
| Vacuum toxin | 981 | 56 | |
| vat-F | TCCTGGGACATAATGGTCAG | ||
| vat-R | GTGTCAGAACGGAATTGT |
3.4. Detection of Class 1 Integrons and Their Gene Cassettes
The E. coli strains were screened for the presence of Class 1 integron elements. Integrase 5' (intI1)-variable region-(qacEΔ1-sul1)3’ were amplified with the primer pair, as shown in Table 1. The identity of the resistance cassettes was analyzed by sequencing performed at the Institute of Biology, UNAM, using a 3730Xl DNA analyzer (Applied Biosystems, Foster City, California, USA) with in-F or in-B primers (Table 1). Nucleotide sequences were compared with the online sequence database (GenBank) using the BlastX algorithm (http://blast.ncbi.nlm.nih.gov). Aeromonas salmonicida 718 with plasmid pRAS1 [IncU, Class 1 integron (intI1-dfrA16qacEΔ1/sul1)] was used as a positive control.
3.5. Detection of Virulence Genes (Virus Typing)
The virulence genes papC, fimH, iutA, cnf1, traT, hlyA, vat, pic, and sat were detected by endpoint PCR (Table 1). The genes papC, fimH, iutA, and traT were amplified by multiplex PCR according to Bravata-Alcantara et al. (11).
3.6. Molecular Typing of Escherichia coli Strains Carrying Identical Genetic Arrays
Strains carrying identical genetic arrangements (gene cassettes) in class 1 integrons were analyzed by Enterobacterial repetitive intergenic consensus (ERIC) PCR to differentiate between single strains and clones (n = 12). The clone identification criteria were based on the dichotomous variable (presence or absence of amplified intergenic regions in isolates with identical gene arrays in the variable region of integrons detected in E. coli strains). The primers used for this purpose are shown in Table 1. PCR conditions were: pre-denaturation for 5 min at 95°C, denaturation for 30 sec at 90°C, annealing for 30 sec at 50°C with the first extension for 1 min at 52°C second, and extension for 1 min at 72°C (30 cycles), with a final extension for 8 min at 72°C. Gene pattern profiles were visually analyzed by intra-gel pattern comparison.
3.7. Statistical Analysis
A chi-square test was used to determine the association between virulence and resistance genes and class 1 integrons in the E. coli strains of vaginal origin classified as commensal and virulent. Fisher's exact test was used for samples with expected frequencies < 5; otherwise, the χ2 test was used. All statistics were performed using SAS software (SAS System, 2020) (12) at a significance level of α = 0.05.
4. Results
4.1. Isolation and Bacterial Identification
This study included one hundred and five E. coli strains from women with cervical dysplasia and monobacterial vaginal infection.
4.2. Antimicrobial Resistance Assay
The highest antimicrobial activity against all the tested strains was observed for nitrofurans, amphenicols, and aminoglycosides. The resistance and sensitivity profiles of all the strains are shown in Figure 1.
The antimicrobial resistance profile of Escherichia coli strains isolated from dysplastic patients with vaginal infection [antibiotics: ampicillin (AM); carbenicillin (CB); cephalothin (CF); cefotaxime (CTX); ciprofloxacin (CIP); norfloxacin (NOR); chloramphenicol (C); nitrofurantoin (FM); amikacin (AN); gentamicin (GM); netilmicin (NET); trimethoprim/sulfamethoxazole (SXT)]. A, β-lactamics; B, quinolones; C, amphenicols; D, nitrofurans; E, aminoglycosides; F, sulfamides.
4.3. Phylogenetic Assignment
The most prevalent phylogenetic group was A/C (38/36.19%), followed by the group B2 (24/22.85%), the group E (17/16.19%), the unknown group (12/11.48%), the group D (7/6.6%), and finally the group F (7/6.6%). The phylogenetic distribution of commensal (the groups A/C and F and the unknown group) and virulent strains (the groups B2, D, and E) was 54.28 and 45.71%, respectively.
4.4. Detection of Class 1 Integrons and Their Gene Cassettes
The integrase gene was amplified by PCR in the strains, of which 44 (41.9%) carried this gene. Once positive amplification against the first molecular target was obtained, a second reaction was performed on the 44 strains against the qacEΔ1-sul1 gene. Finally, a third reaction was used to analyze the gene set of the variable region [flanked by conserved regions (attI1 and qacEΔ1)] in the 44 strains. The absence of PCR products in these strains allowed 28 integrons to be classified as "empty integrons." Sixteen complete integrons (15%) were identified, of which nine were present in commensal strains and seven in virulent strains. The size of variable region amplicons ranged from 474 bp to 1610 bp and showed diverse genetic arrangements in class 1 integrons. The criteria for defining the identity of the obtained arrays were percent match (generally > 75%), match length (generally > 100 bp), and similarity probability.
The sequences showed inserted cassettes corresponding to genes encoding adenylyltransferases (aadA1 and aadA5), dihydrofolate reductases (dfrB4, dfrA1, dfr2D, dfrA7, and dfrA17), and oxacillinases (blaOXA-1). Eight different gene sets were identified in the 16 strains. The blaOXA-1-aadA1 gene set predominated over other gene sets identified in the study (5/16). The aadA5-dfrA17 gene array was present in three strains (22, 32, and 84) and aadA1-dfrA1 in only one (54). The aadA1 gene array, aadA1, was identified in two strains (13 and 27), and dfrB4 was identified in two strains (35 and 48). Individual arrays with dfrA7, dfrA17, and dfr2D cassette variants were identified in three different strains (58, 64, and 102 strains, respectively) (Figure 2). The phylogenetic distribution of the integron-bearing strains (n = 16) was as follows: the group A/C (8/7.6%) was the most predominant, followed by the group D (3/2.8%), the group B2 (2/1.9%), and the group E or clade I (2/1.9%), and finally the unknown group (1/1%). No statistically significant differences were found in the variable region genes (P > 0.05).
Resistance profiles, resistance genotype (class 1 integrons), virus typing, and Clermont genotyping in Escherichia coli strains isolated from dysplastic patients with vaginal infection. The positivity for a given phenotype or genotype is depicted with a black rectangle (the numbers 1 to 8 were used to indicate gene cassettes: 1, aadA1; 2, aadA5; 3, dfrA1; 4, dfr2D; 5, dfrB4; 6, dfrA7; 7, dfrA17; 8, blaOXA-1).
4.5. Detection of Virulence Genes
The virulence genes identified in the E. coli strains were fimH (94/89.5%), traT (74/70.4%), iutA (72/68.5%), papC (42/40%), pic (13/12.3%), sat (7/6.6%), cnf1 (5/4.7%), hlyA (3/2.8%), and vat (2/1.9%) (Figure 2). Significant differences (P < 0.05) were only identified in the traT and pic genes belonging to the commensal strains (group A / C) when they were compared with the virulent strains.
4.6. Relationship Between Properties of Virulence Genes
Adhesion genes (fimH and papC) were detected, followed by phagocytosis evasion (traT) and iron uptake (iutA) genes. The percentage of cytotoxic genes was low. Genetic associations between the fimH, iutA, and traT genes were the most predominant in commensal and virulent strains. The frequency of the virulence genes fimH, iutA, and traT was higher in the phylogenetic groups B2 of virulent strains and A/C of commensal strains. Cytotoxic genes showed the lowest incidence in both commensal and virulent strains. There were no other significant genetic associations with the other virulence genes. As shown in Table 2, no significant differences were observed between the genetic associations of these genes (P > 0.05). The table also shows the relationship between the properties of virulence genes in the E. coli strains.
| Phylogenetic Groups | Virulence Gene; No. (%) | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Adherence | Cytotoxic | Siderophore | Phagocytosis Evasion | ||||||
| fimH | papC | hlyA | cnf1 | sat | pic | vat | IutA | traT | |
| Virulent | |||||||||
| B2 (24) | 23 (95.8) | 10 (41.6) | 1 (4.1) | 1 (4.1) | 4 (16.6) | 1 (4.1) | 1 (4.1) | 20 (83.3) | 18 (75) |
| D (13) | 12 (92.3) | 8 (61.5) | 1 (7.7) | 0 | 0 | 1 (7.7) | 0 | 8 (61.5) | 8 (61.5) |
| E (11) | 10 (90.9) | 2 (18.2) | 0 | 0 | 0 | 0 | 0 | 5 (45.5) | 8 (72.7) |
| Commensal | |||||||||
| A/C (38) | 35 (92.1) | 16 (42.1) | 0 | 3 (7.89) | 2 (5.3) | 7 (18.5) | 1 (2.6) | 28 (73.7) | 28 (73.7) |
| F (7) | 5 (71.4) | 3 (42.9) | 1 (14.3) | 1 (14.3) | 1 (14.3) | 1 (14.3) | 0 | 4 (57.1) | 2 (28.6) |
| Unknown (11) | 9 (81.8) | 3 (27.3) | 0 | 0 | 0 | 3 (27.3) | 0 | 7 (63.6) | 10 (90.9) |
4.7. Molecular Typing of Escherichia coli Strains Carrying Identical Genetic Cassettes Arrays
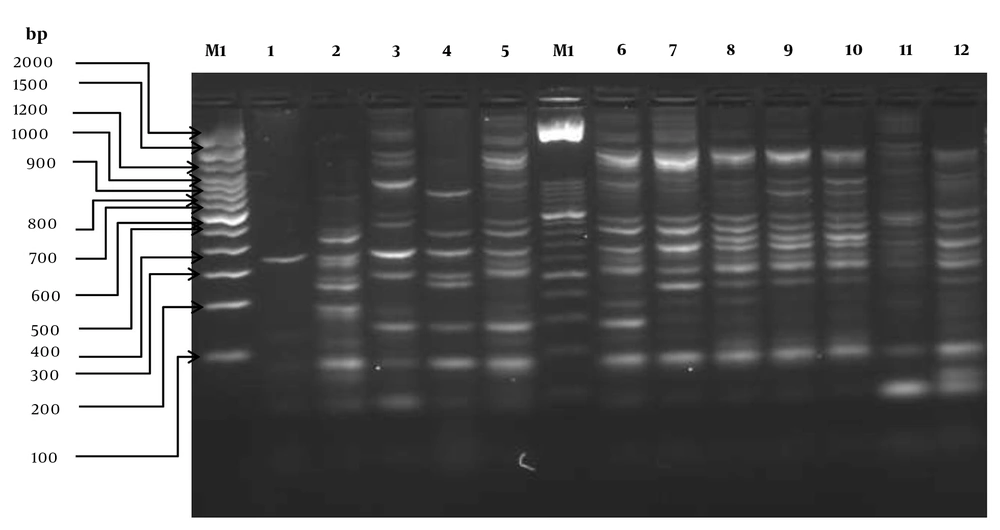
PCR product analysis and gel electrophoresis showed three to 16 bands in different profiles. Several amplicons were found, ranging in size from ≈150 bp to about ≈2000 bp. DNA fragments between 200 - 400 bp were highly frequent. Diversity in the identification of intergenic regions allowed us to differentiate 15 unique strains. This probe showed that 14 of the 16 E. coli strains were unique and were not clones (Figure 3). Only two strains (60 and 76) were indistinguishable from each other (100% similarity) and carried the same gene set (blaOXA-1/aadA1). Figure 2 shows the clonal distribution obtained by ERIC-PCR among E. coli strains carrying class 1 integrons and their phylogenetic relationship by the Clermont algorithm.
Electrophoretic patterns generated by the ERIC-PCR of Escherichia coli strains isolated from dysplasic patients with vaginal infection [lanes: M1, 100bp molecular size marker (Invitrogen TM); M2, 50bp molecular size marker (Invitrogen TM); 1 – 2, aadA1, 3 – 5, aadA5/dfrA17, 6-10:blaOXA-1/aadA1, 11-12: dfrB4. Identical cassette arrays: line 9 and 10 (blaOXA-1/aadA1).
5. Discussion
In this study, the E. coli strains showed resistance mainly to β-lactams and sulfonamides; similar results were observed in a previous study (7). Seventy-three percent of the strains were resistant to three or more antibiotics, agreeing with other reports (13, 14). Resistance to these antibiotics has previously been observed due to antibiotic efflux pumps (15) or class 1 integrons that may carry genes conferring resistance to different antibiotic families (7). In most of the sulfonamide-resistant strains, the sul1 gene was present (16). Some of the isolates showed susceptibility to the corresponding antibiotic, and others were resistant. This may be attributed to the loss of expression of integron-localized genes (17) or the fact that these genes were located in regions other than integrons, such as in chromosomes or plasmids (6). However, E. coli isolates with integrons were more resistant to all the tested antibiotics than those without integrons. The identification of empty integrons in a high proportion in this work indicates the potential to capture antibiotic-resistant cassettes. The integrons harbored major antibiotic resistance genes used in treating urinary tract infections, such as dfr, which encodes variants of the enzyme dihydrofolate reductase and provides resistance to trimethoprim (18).
In Mexico, trimethoprim has been frequently used as the antibiotic of choice for treating urinary tract infections, which has led to recurrent vaginal infections associated with the presence of the dfrA genes (19). The blaOXA-1 gene conferring resistance to β-lactams (6), variants of the addA gene for streptomycin, and the sul1 gene for sulfonamides (19) were also found. The highest incidence of monogenic arrays found in this work was constituted by the dfr genes, as in other studies (20). Trimethoprim-resistant strains have previously been reported to be highly resistant to other antibiotics (18). In this study, we found high resistance to ampicillin and carbenicillin in trimethoprim-resistant E. coli strains. In addition, there may be a correlation between integron-bearing strains and resistance to a higher number of antibiotics (5).
The genomic capacity of E. coli allows the expression of numerous virulence factors responsible for infection (4). The function of the fimH gene product is essential in the initial phase of infection by recognizing receptors on the host cell surface that facilitate bacterial colonization (21). In addition, this result could be related to the pathogenicity of the isolated strains (4). The fimH, iutA, and traT genes were most frequently found in the phylogenetic group B2, which has been reported to be a virulent group (22). Interestingly, the incidence of these genes was high in the phylogenetic group A/C, indicating that most strains were related to three critical virulent properties, including adhesion (fimH), evasion of phagocytosis (traT), and iron uptake (iutA), and might be involved in the development of infection; similar virulence gene profiles were previously described (23). The expression of other adhesins, such as the pap gene, may confer advantages to E. coli strains for ascent and pathogenesis (24). Similar to this work, Oliveira et al. (25) reported the presence of at least one of these virulence genes in strains of E. coli (traT, papC, cnf1, and hlyA).
This shows that the isolates, even when classified as commensal strains, carry virulence factors necessary to cause an infectious process. Also, depending on the amount and virulence of the combination of present genes, the pathogenic potential of the E. coli bacterium will depend on the amount and appropriate virulence of the combination of present genes (26). It is interesting to know that even when genotyping by Clermont's algorithm provides information on the virulence role of the strains, a large number of these strains have genes associated with essential virulence factors, such as those mentioned above. The information shown in this work sets guidelines to reconsider this genotyping method as a standard protocol for classifying assemblages according to their virulence and commensalism.
We hypothesize that the genetic background identified in all strains results from the constant evolutionary change of this microorganism, which confers its properties to recognize and colonize new ecological niches in human mucosa and epithelia. In our work, we observed a correlation between the presence of the fimH, traT, iutA, and papC genes with high resistance to β-lactams and sulfonamides. Future work will be directed to study the expression levels of these genes by using in vitro infection models of cell lines, as previously reported (19, 21). The findings showed that even when the presence of virulence genes is high, the expression of these genes could be variable in in vivo models. This highlights the importance of the study of gene expression to give more support to the results obtained in this work.
5.1. Conclusions
The high incidence of virulence and resistance factors in commensal and virulent strains of E. coli revealed potential tools in the pathogenesis of vaginal infection in dysplastic patients and determined a significant detection challenge for the clinical microbiology laboratory.

![The antimicrobial resistance profile of <i>Escherichia coli</i> strains isolated from dysplastic patients with vaginal infection [antibiotics: ampicillin (AM); carbenicillin (CB); cephalothin (CF); cefotaxime (CTX); ciprofloxacin (CIP); norfloxacin (NOR); chloramphenicol (C); nitrofurantoin (FM); amikacin (AN); gentamicin (GM); netilmicin (NET); trimethoprim/sulfamethoxazole (SXT)]. A, β-lactamics; B, quinolones; C, amphenicols; D, nitrofurans; E, aminoglycosides; F, sulfamides. The antimicrobial resistance profile of <i>Escherichia coli</i> strains isolated from dysplastic patients with vaginal infection [antibiotics: ampicillin (AM); carbenicillin (CB); cephalothin (CF); cefotaxime (CTX); ciprofloxacin (CIP); norfloxacin (NOR); chloramphenicol (C); nitrofurantoin (FM); amikacin (AN); gentamicin (GM); netilmicin (NET); trimethoprim/sulfamethoxazole (SXT)]. A, β-lactamics; B, quinolones; C, amphenicols; D, nitrofurans; E, aminoglycosides; F, sulfamides.](https://services.brieflands.com/cdn/serve/3170b/1f5b036015d5ae9aaf5702fad38c1cc1dbb79797/jjm-117488-i001-F1-preview.webp)