Abstract
Background:
Uropathogenic Escherichia coli (UPEC) strains, encoding superficial and secretory virulence factors, can lead to colonization and facilitation of bacterial growth in the host urinary tract, causing Urinary Tract Infection (UTI).Objectives:
This study determined the ability of biofilm formation by the Congo red agar method, the presence of virulence genes using the multiplex polymerase chain reaction (PCR) method, and the relationship between biofilm formation and antibiotic resistance patterns and virulence genes in E. coli clinical isolates in Yasuj.Methods:
This cross-sectional study was performed on 144 UPEC isolates collected in 2017. Biofilm formation was detected by the Congo red agar phenotypic assay and virulence factors by the multiplex PCR method. Antibiotic resistance tests were performed by the Kirby-Bauer method.Results:
Out of 144 isolates of E. coli, 22 (19.4%) isolates showed to be strong biofilm producers, 27 (23.8%) moderate biofilm producers, and 64 (56.3%) weak biofilm producers. A significant relationship was observed between biofilm-producing strains and resistance to ampicillin (P = 0.020) and cotrimoxazole (P = 0.038). The virulence genes in strong biofilm producers included iutA (95%), FimH (93%), ompT (90%), PAI (90%), and TraT (81%) genes. The phylogroup B2 carried the most virulence genes. A significant correlation was observed between E. coli phylogenetic groups and aer (P = 0.019), iroN (P = 0.042), and ompT (P = 0.032) virulence genes.Conclusions:
The results of this study showed a high prevalence of virulence genes, and antibiotic-resistant E. coli strains capable of biofilm formation. The results of this study may help elucidate the pathogenesis of UPEC and facilitate better treatment strategies for patients with UTIs in this geographic area.Keywords
Uropathogenic Escherichia coli Virulence Factors Drug Resistance Microbial Phylogenetic Groups Urinary Tract Infections
1. Background
Urinary tract infection (UTI) is one of the most common human infections of bacterial origin. Among the bacteria that cause UTIs, the strains of E. coli, called uropathogenic Escherichia coli (UPEC), are the most important causes of this infection. This infection is one of the main causes of hospitalization with significant complications and high healthcare costs (1, 2). Today, E. coli phylogenetic groups are determined based on the presence of chuA, TspE4.C2, and yjaA genes. According to the results of various studies, most uropathogenic E. coli strains are in the B2 and D phylogroups while commensal strains belong to groups A and B1 (3). The ability of E. coli strains to cause urinary tract infections is due to the formation of biofilms and the presence of many virulence factors that depend on the invasion, colonization, and survival of uroepithelium cells (4, 5). Bacteria with these factors can fight host defense factors such as cytokines, including interleukin-8, urinary flow, and Tamm-Horsfall proteins (uroepithelial cell defensin peptides). Therefore, the pathogenicity of E. coli strains in UTIs depends on the balance between the host and these bacterial virulence factors (6).
Biofilms are microbial communities enclosed in the extracellular polymeric matrix composed of nucleic acids, proteins, and enzymes that bind to living and non-living surfaces. Biofilms can increase the survival of bacteria in the urinary tract by protecting them against the cleansing effects of hydrodynamic forces, host defense mechanisms, phagocytosis, and antibiotics. Therefore, biofilm production plays an important role in the pathogenicity of UPEC strains (7-10). Uropathogenic E. coli strains are pathogenic due to virulence factors such as adhesion fimbriae (fim-H, iha), toxins (cnf1, hlyA), iron-forming systems (iroN, aer), macrophage degradation agents (ompT protease), and serum resistance factors (traT), which are commonly encoded in Pathogenicity Islands (PAI). Besides, serum resistance factors (traT) contribute to the pathogenesis of E. coli strains in UTIs (11-14).
The pathogenicity islands (PAIs) are specific regions on the bacterial chromosome where virulence genes accumulate. PAIs and their associated virulence genes spread among bacterial populations by horizontal transfer (8). Increased antibiotic resistance (due to overuse and improper use of antibiotics) among pathogens, especially those causing UTIs, is a major problem that is the main reason for the emergence of resistant strains (especially multidrug-resistant strains), dissemination of resistance factors to susceptible strains, increased treatment costs, treatment failure, and death (3, 14, 15). Due to the increasing infections associated with E. coli and different factors involved in bacterial pathogenesis in different parts of the world, as well as the emergence of drug-resistant strains, it seems necessary to study pathogenic factors in drug-resistant bacteria (16).
2. Objectives
This study determined the ability of biofilm formation by the Congo red agar method, the presence of virulence genes and phylogenetic groups using the multiplex PCR method, and the relationship between biofilm formation and antibiotic resistance patterns and virulence genes in E. coli clinical isolates in Yasuj.
3. Methods
In this cross-sectional study, 144 E. coli isolates were collected from urine samples of patients with UTIs who had been referred to Imam Sajjad and Shahid Beheshti hospitals in Yasuj. The isolates were incubated on blood agar and Eosin methylene blue agar (Merck, Germany) at 37°C for 24 h. After culture, they were identified and confirmed using standard microbiological and biochemical methods. The biochemical analyses like indole test, citrate test, triple sugar iron agar test, and urease test were performed for the identification of microorganisms. The isolates were finally stored in Trypticase soy broth (Merck, Germany), with 20% glycerol at -20°C for the study of biofilm formation and DNA extraction (3).
3.1. Biofilm Production Assays
Biofilm production was detected using the Congo red agar method, following the procedure described by Freeman et al. The Congo red agar medium was prepared by mixing its ingredients such as agar (Merck, Germany), sucrose, and Congo red dye and brain-heart infusion broth in one liter of distilled water. The Congo red agar reagent was prepared separately as an aqueous solution and autoclaved at 121°C for 10 min. After preparation of brain-heart infusion broth with agar 10 g/mL, the reagent with 50 g/L sucrose was added to the medium. By removing a single bacterial colony from the Eosin methylene blue agar medium (Merck, Germany) with a loop, it was cultured on the surface of the Congo red agar medium. Finally, the plates were incubated for 24 to 48 h at 37°C. Biofilm-producing strains were divided into three groups: strong biofilm producers, moderate biofilm producers, and weak biofilm producers (17, 18).
3.2. Antimicrobial Susceptibility Test
We assessed the susceptibility of E. coli isolates to the antibiotics ciprofloxacin (30 μg), nalidixic acid (30 μg), cefotaxime (30 μg), cotrimoxazole (25 μg), ampicillin (10 μg), ceftriaxone (30 μg), tetracycline (30 μg), aztreonam (30 μg), ceftizoxime (30 μg), and amikacin (30 μg; BD-BBL Company, America) using the Kirby-Bauer disk diffusion method as per the CLSI and clinical standards. To control the quality of the disks, E. coli ATCC 25922 was used (3).
3.3. DNA Extraction
In the present study, genomic DNA extraction was performed using the boiling method. Briefly, several colonies of bacteria (24 h) were first dissolved in 300 μL of distilled water. It was then boiled at 95°C for 10 min. Centrifugation was done at 12,000 rpm for 10 min. To perform the PCR method, the supernatant was stored as a template DNA (9).
3.4. PCR Method to Determine Virulence Genes and Phylogenetic Groups
All isolates were tested for the presence of virulence genes such as fimH, ihA, iroN, iutA, aer, ompT, traT, Pai, cnf1, and hlyA genes. The primers used to detect the virulence genes in this study are shown in Table 1, which were purchased from Pishgam Company (Iran). In the present study, the phylogenetic groups of E. coli isolates were determined using triplex PCR with chuA, TspE4.C2, and yjaA primers as described by Clermont et al. (19). The PCR program to identify the virulence genes and phylogenetic groups of E. coli strains was as follows: (1) initial denaturation at 94°C for 5 min; (2) 30 cycles including denaturation at 94°C for one minute; and (3) binding the primers (annealing) of the phylogenetic groups to the template DNA at 59°C for 20 s. The binding temperatures of virulence gene primers are listed in Table 1. The extension stage was done at 72°C for one minute, and the final extension stage was performed at 72°C for 10 min (Table 1). Electrophoresis of PCR products was performed on a 2% agarose gel with DNA safe stain dye solution in the presence of a 100 bp marker (Pishgam, Iran) and 90-volt constant voltage for 65 min. The gel was then examined with a UV Transilluminator (Major Science, Taiwan) (Figures 1 and 2).
Primers Used to Detect Virulence Genes in Uropathogenic Escherichia coli Isolates
| Gens | Sequences (5′ → 3′) | Product size (bp) | Annealing Tm, (°C) | References |
|---|---|---|---|---|
| FimH | 400 | 55 | (20) | |
| Forward | GTTGTTCTGTCGGCTCTGTC | |||
| Reverse | TAAATGTCGCACCATCCAG | |||
| ihA | 827 | 58 | (21) | |
| Forward | CTGGCGGAGGCTCTGAGATCA | |||
| Reverse | TCCTTAAGCTCCCGCGGCTGA | |||
| iroN | 1048 | 58 | (22) | |
| Forward | CGGTTCCTGGCACGAATATCAT | |||
| Reverse | TTTTGGGATTTCCCCAACCTGG | |||
| iutA | 300 | 63 | (23) | |
| Forward | GGCTGGACATCATGGGAACTGG | |||
| Reverse | CGTCGGGAACGGGTAGAATCG | |||
| aer | 602 | 61 | (24) | |
| Forward | TACCGGATTGTCATATGCAGACCGT | |||
| Reverse | AATATCTTCCTCCAGTCCGGAGAAG | |||
| ompT | 559 | 58 | (25) | |
| Forward | ATCTAGCCGAAGAAGGAGGC | |||
| Reverse | CCCGGGTCATAGTGTTCATC | |||
| TraT | 290 | 63 | (24) | |
| Forward | GGTGTGGTGCGATGAGCACAG | |||
| Reverse | CACGGTTCAGCCATCCCTGAG | |||
| Cnf1 | 498 | 63 | (24) | |
| Forward | AAGATGGAGTTTCCTATGCAGGAG | |||
| Reverse | CATTCAGAGTCCTGCCCTCATTATT | |||
| HlyA | 1177 | 63 | (26) | |
| Forward | AACAAGGATAAGCACTGTTCTGGCT | |||
| Reverse | ACCATATAAGCGGTCATTCCCGTCA | |||
| PAI | 930 | 63 | (24) | |
| Forward | GGACATCCTGTTACAGCGCGCA | |||
| Reverse | TCGCCACCAATCACAGCCGAAC |
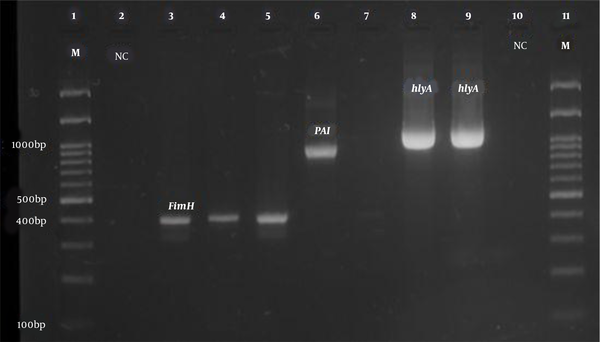
PCR product electrophoresis for FimH, PAI, and hlyA genes (Row M, marker 100 bp DNA ladder; Rows 3, 4, and 5, positive sample of FimH gene 400 bp; Row 6, positive sample of PAI gene 930 bp; Rows 8 and 9, positive sample of hlyA gene 1177 bp; Rows 2 and 10, negative control).
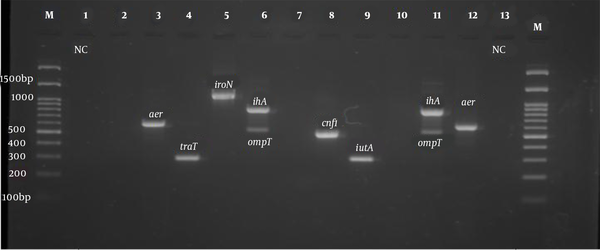
PCR product electrophoresis for aer, iroN, ompT, ihA, and cnf1 genes (Row M, marker 100 bp DNA ladder; Rows 3 and 12, positive sample of aer 602 bp; Row 4, positive sample of TraT gene 290 bp; Row 5, positive sample of iroN gene 1148 bp; Rows 6 and 11, positive sample of ihA 857 bp; Rows 6 and 11, positive sample of ompT gene 559 bp; Row 8, positive sample of cnf1 gene 498 bp, and Rows 1 and 13, negative control).
3.5. Statistical Analysis
We used SPSS version 18.0 software for statistical analysis. Fisher's exact and chi-square tests were used to investigate the relationship between the formation of biofilms and virulence genes. The significance level was set at P-value < 0.05.
4. Results
4.1. Distribution of Virulence Genes
According to the results of the PCR test for the identification of surveyed virulence genes, the highest frequency belonged to the FimH gene, which was detected in 93.8% of the isolates. The prevalence of iutA virulence gene was 90.3% (130 samples), traT 88.8% (128 samples), ompT 88.2% (127 samples), PAI 77.8% (112 samples), aer 62.5% (90 Sample), ihA 59.7% (86 samples), cnf1 toxin-related gene 41.7% (60 samples), and HlyA 34.7% (50 samples). The iroN gene had the lowest frequency as 32.6% (47 samples). The expression patterns of virulence genes are shown in Table 2.
Results on the Frequency of Virulence Genes and Selected Patterns of Urinary Tract Infection
| Type of Virulence Factors | Virulence Genes | No. (%) (n = 144) |
|---|---|---|
| Adhesion | fimH | 135 (93.8) |
| ihA | 86 (59.7) | |
| Iron acquisition systems | iutA | 130 (90.3) |
| iroN | 47 (32.6) | |
| aer | 90 (62.5) | |
| Toxins | Cnf1 | 60 (41.7) |
| HlyA | 50 (34.7) | |
| Outer membrane proteins | ompT | 127 (88.2) |
| Serum Resistance | traT | 128 (88.8) |
| pathogenicity islands | PAI | 112 (77.8) |
| Patterns of Gene Expression | ||
| Pattern Codes | Virulence Genes | No. (%) (n = 144) |
| E1 | FimH, ihA | 81 (56.25) |
| E2 | FimH, iutA | 122 (84.72) |
| E3 | IutA, iron | 42 (29.16) |
| E4 | IroN, aer | 20 (13.8) |
| E5 | ompT, traT | 112 (77.77) |
| E6 | OmpT, PAI | 99 (68.75) |
| E7 | PAI.HlyA | 39 (27.08) |
| E8 | traT, cnf1 | 53 (36.8) |
| E9 | HlyA, cnf1 | 27 (18.75) |
| E10 | PAI, ompT, traT | 91 (63.19) |
| E11 | PAI, cnf1, hlyA | 20 (13.8) |
| E12 | HlyA, cnf1, ompT, traT | 23 (15.97) |
| E13 | fimH, ompT, iuta, PAI | 86 (59.72) |
| E14 | FimH, PAI, ompT, traT | 87 (60.41) |
| E15 | FimH, iha, HlyA, cnf1 | 17 (11.80) |
| E16 | ompT, traT, fimH, iha, HlyA, cnf1 | 14 (9.7) |
| E17 | ompT, traT, fimH, iha, HlyA, cnf1, cnf1 | 9 (6.25) |
The Triplex PCR results showed that 106 (73.6%) isolates belonged to the phylogenetic group B2, 23 (15.9%) isolates belonged to group D, eight (5.5%) isolates belonged to group B1, and seven (4.86%) isolates belonged to group A. Also, the distribution of virulence genes among the phylogenetic groups of E. coli was investigated that showed the prevalence of virulence genes belonging to group B2 was higher than those of other phylogenetic groups. The highest frequency of pathogenic genes in group B2 belonged to fimH (94%), iutA (92%), ompT (90%), traT (89%), PAI (76%), aer (69%), ihA (62%), cnf1 (41%), hlyA (34%), and iroN (31%). A significant correlation was observed between E. coli phylogenetic groups and aer (P = 0.019), iroN (P = 0.042), and ompT (P = 0.032) virulence genes (Table 3).
Distribution of Virulence Genes Among Phylogenetic Groups of Uropathogenic Escherichia coli a
| Phylogenetic Group | No. Strains | Virulence Factors | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| fimH | iutA | ihA | PAI | aer | iroN | TraT | hlyA | CNF | ompT | ||
| A | 7 (4.8) | 6 (85) | 5 (71) | 5 (71) | 5 (71) | 2 (28) | 3 (31) | 6 (85) | 2 (28) | 3 (42) | 4 (51) |
| B1 | 8 (5.6) | 7 (87) | 6 (755) | 2 (25) | 5 (62) | 4 (50) | 3 (42) | 5 (62) | 2 (25) | 5 (62) | 8 (100) |
| B2 | 106 (73.6) | 100 (94) | 98 (92) | 66 (62) | 81 (76) | 74 (69) | 33 (31) | 95 (89) | 36 (34) | 44 (41) | 96 (90) |
| D | 23 (16) | 22 (95) | 21 (91) | 13 (25) | 21 (91) | 10 (43) | 5 (21) | 22 (95) | 10 (43) | 8 (34) | 19 (82) |
| P-value | 0.680 | 0.135 | 0.188 | 0.285 | 0.019 | 0.042 | 0.077 | 0.740 | 0.597 | 0.032 | |
4.2. Results of Biofilm Formation in Escherichia coli Strains
Based on the Congo red agar method for biofilm formation, out of 144 E. coli isolates from UTI cases in Yasuj, 113 isolates could form biofilms. The results were interpreted based on the appearance of the colony. Out of 113 isolates with positive biofilms, 22 isolates had very black colonies (strong biofilm producers), 27 isolates had black colonies with smooth and round surfaces (medium biofilm producers), and 64 isolates had gray colonies (weak biofilm producers). Biofilm production was significantly associated with cnf1 virulence genes (P = 0.038), but no significant association was observed with other virulence genes (Table 4). In this study, the expression of virulence genes was investigated by the PCR method. The prevalence of virulence genes in biofilm-producing strains was as follows. The results showed that among biofilm producers, the prevalence of fimH, iutA, ompT, traT, PAI, aer, ihA, Cnf1, hlyA, and iroN genes was 93.8, 92, 89, 89, 79, 62, 61, 46, 35, and 29%, respectively (Table 4).
The biofilm forming isolates showed maximum resistance to ampicillin (85%), tetracycline (69%), cotrimoxazole (66%), ceftizoxime (64%), aztreonam (61%), nalidixic acid (59%), ceftriaxone (53%), ciprofloxacin (52%), cefotaxime (50%) and the lowest to amikacin (0%). In this study, a significant relationship was observed between the strains of biofilm formation and resistance to ampicillin (P = 0.020) and cotrimoxazole (P = 0.038) antibiotics (Table 5). Besides, we investigated the relationship between the presence of virulence genes in E. coli strains and antibiotic resistance patterns, as shown in Table 6.
Frequency of Virulence Genes in Escherichia coli Isolates of Uropathogen-Forming Biofilms a
| Gens Virulence | No. (%) of Strains | P-Value | |||
|---|---|---|---|---|---|
| Strong Biofilm Producers (n = 22 ) | Moderate Biofilm Producers (n = 27) | Weak Biofilm Producers (n = 64) | Total | ||
| FimH | 20 (93.8) | 25 (92.6) | 61 (95.3) | 106 (93.8) | 0.728 |
| ihA | 14 (63.3) | 14 (51.9) | 42 (65.6) | 70 (61.9) | 0.458 |
| Hly | 11 (50) | 8 (29.6) | 21 (32.8) | 40 (35.4) | 0.268 |
| Cnf1 | 15 (68.2) | 14 (51.9) | 24 (37.5) | 53 (46.9) | 0.038 |
| iroN | 8 (36.4) | 8 (29.65) | 17 (26.6) | 33 (29.2) | 0.683 |
| iutA | 21 (95.5) | 25 (92.6) | 58 (90.6) | 104 (92) | 0.765 |
| Aer | 13 (59.1) | 20 (74.1) | 38 (59.4) | 71 (62.8) | 0.383 |
| ompT | 20 (90.9) | 25 (92.6) | 56 (87.5) | 101 (89.4) | 0.746 |
| TraT | 18 (81.8) | 23 (85.8) | 60 (93.8) | 101 (89.4) | 0.211 |
| PAI | 20 (90.9) | 20 (74.1) | 50 (78.1) | 90 (79.6) | 0.312 |
Antibiotic Susceptibility Results of Biofilm and Non-biofilm Producing Uropathogenic Escherichia coli by Congo Red Agar
| Antibiotics | Biofilm Producers; 113 (78.4%) | Non-biofilm Producers; 31 (21.5%) | P-Value | ||
|---|---|---|---|---|---|
| R | S | R | S | ||
| Ampicillin | 97 (85) | 16 (14) | 21 (67) | 10 (32) | 0.020 a |
| Cefotaxime | 57 (50) | 53 (46) | 12 (38) | 19 (61) | 0.611 |
| Ceftriaxone | 61 (53) | 52 (46) | 12 (38) | 19 (61) | 0.406 |
| Ceftizoxime | 73 (64) | 40 (35) | 11 (35) | 20 (64) | 0.109 |
| Cotrimoxazole | 75 (66) | 37 (32.7) | 16 (51) | 15 (48) | 0.038 a |
| Aztreonam | 69 (61) | 34 (30) | 18 (58) | 13 (41) | 0.622 |
| ciprofloxacin | 59 (52) | 54 (47.7) | 14 (45) | 17 (54) | 0.422 |
| Nalidixic acid | 67 (59) | 46 (40) | 15 (48) | 16 (51) | 0.093 |
| Tetracycline | 78 (69) | 35 (30.9) | 18 (58) | 13 (41) | 0.325 |
Distribution of Virulence Genes in Antibiotic Resistance Patterns of Escherichia coli Isolates
| Virulence Gen | Antibiotic | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Ampicillin | Cefotaxime | Ceftriaxone | Ceftizoxime | Cotrimoxazole | Aztreonam | Ciprofloxacin | Nalidixic Acid | tetracycline | |
| FimH | 109 (80) | 56 (41) | 56 (41) | 57 (42) | 77(54) | 73(54) | 45 (33) | 63 (46) | 83 (63) |
| P-value | 0.889 | 0.552 | 0.538 | 0.258 | 0.167 | 0.316 | 0.100 | 0.410 | 0.249 |
| ihA | 71 (82) | 42 (48) | 41 (47) | 43 (50) | 54(62) | 55(64) | 41 (47) | 45 (52) | 52 (67) |
| P-value | 0.757 | 0.137 | 0.277 | 0.099 | 0.528 | 0.037 | 0.001 | 0.185 | 0.971 |
| iutA | 105 (80) | 57 (43) | 54 (41) | 58 (44) | 79 (60) | 74 (56) | 46 (35) | 63 (48) | 77 (59) |
| P-value | 0.852 | 0.417 | 0.831 | 0.738 | 0.156 | 0.425 | 0.994 | 0.757 | 0.561 |
| iroN | 37 (78) | 14 (29) | 9 (19) | 16 (34) | 26 (55) | 24 (51) | 12 (25) | 20 (42) | 28 (59) |
| P-value | 0.497 | 0.027 | 0.000 | 0.263 | 0.745 | 0.601 | 0.181 | 0.655 | 0.241 |
| aer | 69 (76) | 42 (46) | 44 (48) | 46 (51) | 51 (56) | 53 (58) | 35 (38) | 50 (72) | 52 (57) |
| P-value | 0.236 | 0.032 | 0.29 | 0.005 | 0.662 | 0.149 | 0.069 | 0.051 | 0.445 |
| Hly A | 39 (78) | 14 (28) | 18 (36%) | 16 (32) | 29 (58) | 21 (42) | 11 (22) | 23 (46) | 33 (66) |
| P-value | 0.148 | 0.034 | 0.148 | 0.106 | 0.902 | 0.043 | 0.032 | 0.530 | 0.394 |
| CNF | 45 (75) | 21 (35) | 21 (35) | 23 (38) | 35 (58) | 33 (55) | 22 (36) | 26 (43) | 23 (38) |
| P-value | 0.140 | 0.137 | 0.186 | 0.387 | 0.725 | 0.537 | 0.323 | 0.176 | 0.330 |
| TraT | 104 (81) | 56 (43) | 56 (43) | 55 (44) | 76 (59) | 75 (58) | 48 (37) | 62 (48) | 78 (60) |
| P-value | 0.670 | 0.205 | 0.182 | 0.380 | 0.552 | 0.116 | 0.237 | 0.298 | 0.380 |
| ompT | 103 (81) | 52 (40) | 56 (40) | 53 (41) | 74 (58) | 75 (58) | 44 (34) | 64 (50) | 78 (61) |
| P-value | 0.241 | 0.642 | 0.344 | 0.487 | 0.824 | 0.722 | 0.176 | 0.266 | 0.335 |
| PAI | 90 (80) | 51 (45) | 52 (46) | 51 (45) | 64 (57) | 65 (58) | 42 (37) | 59 (52) | 68 (60) |
| P-value | 0.598 | 0.274 | 0.487 | 0.717 | 0.311 | 0.311 | 0.587 | 0.007 | 0.858 |
5. Discussion
As known, E. coli is the cause of 80 - 90% of community-acquired UTIs and 30 - 50% of nosocomial UTIs (27). One of the important factors involved in the pathogenesis of E. coli is the production of biofilms. Biofilm formation in E. coli can contribute to bacterial adhesion and colonization in the host urinary tract. Bacteria cause many infections by forming biofilms, which are difficult to treat because they increase resistance to antibiotics by producing biofilms (28-30). In our study, Congo red agar biofilm formation was observed in 78% of the isolates, which was also studied by Poursina et al. (80%), Katongole et al. (78%), Sudheendra and Basavaraj (71%), Poovendran and Ramanathan (79%), and Neupane et al. (69%) (16, 31-34). Besides, Tajbakhsh et al. (61%) and Niveditha et al. (56%) reported a lower prevalence of biofilm formation, and Ponnusamy et al. reported a 100% prevalence of biofilm formation (5, 35, 36). Differences in biofilm formation can be due to reasons such as the low level of hygiene, differences in geographical areas, study time, increased antibiotic resistance, and differences in the sources of sample isolation.
Examining the effective factors in the formation of bacterial biofilms can help treat infections caused by bacteria (33, 37). Various studies have shown that the bacteria that make up the biofilm protect the bacteria against the penetration of antibiotics due to the structure of the extracellular polymer matrix. In our study, isolates associated with biofilm formation had significant resistance to antibiotics used, such as ampicillin (85%), tetracycline (69%), cotrimoxazole (66%), ceftizoxime (64%), ceftriaxone (53%), aztreonam (61%), nalidixic acid (59%), ciprofloxacin (52%), cefotaxime (50%), and amikacin (0%). The results of our study showed that E. coli isolates with biofilm production ability were associated with increased resistance to various antibiotics, which is consistent with studies conducted in different parts of Iran and the world (5, 38, 39). It seems that the arbitrary use of antibiotics without a doctor's prescription and the availability and the absence of any law prohibiting the use of over-the-counter antibiotics in patients can be important factors in antibiotic resistance in different communities. These findings underscore the need to regulate the use of antimicrobials and institutionalization of antimicrobial stewardship programs in hospitals to limit the spread of resistant microorganisms (40, 41).
Extraintestinal pathogenic E. coli (ExPEC) isolates contain virulence factors such as fimbriae, toxins, iroN acquisition systems, and invasive factors that provide conditions for bacterial pathogenicity. The identification of UPEC virulence genes can be helpful in our understanding of the pathogenicity of the bacterium and minimizing the complications of bacterial infections. There have been many studies on genes involved in E. coli pathogenesis (42).
In this study, the prevalence of viral genes involved in UPEC was investigated by the PCR method. The highest prevalence of virulence genes was related to the fimbriae fimH virulence gene with 93% frequency and iutA virulence gene with 90% frequency, followed by traT (88.9%), ompT (88.2%), PAI (77.8%), aer (62.5%), iha (59.7%), cnf1 (41.7%), hlyA (34.7%) virulence genes, while iroN had the lowest prevalence with a frequency of 32.6%. The results of this study are similar to other previous studies (20, 21, 23, 43-45). Our study results indicated that E. coli strains with a high prevalence of bacterial virulence factors can be major causative agents for UTIs in humans in Yasuj (Iran). Given the prevalence of virulence genes in previous studies, it can be concluded that the variation in virulence genes of E. coli is due to differences in the isolation of UPEC strains in different geographical regions and the difference in the number of samples (9). The relationship between virulence genes and antibiotic resistance is complex. In this study, FimH, iha, aer, PAI, and iutA genes were observed in more-resistant isolates, and a significant relationship was observed between some virulence genes and antibiotics ceftriaxone, cefotaxime, aztreonam, ciprofloxacin, and nalidixic acid, which was consistent with previous studies (46, 47) (Table 6).
In this study, the prevalence of virulence genes in biofilm-forming strains was determined. Our findings were consistent with the results of previous studies (26, 38, 48, 49). It seems that examining the effective factors in the formation of bacterial biofilms can help treat infections caused by bacteria. We found that E. coli isolates capable of forming strong-to-moderate biofilms had a high prevalence of virulence genes, which may indicate the role of virulence factors in the development of biofilm formation. In our study, a significant relationship was observed between the strains of biofilm formation and resistance to ampicillin (P = 0.020) and cotrimoxazole (P = 0.038) antibiotics. In this study, biofilm formation was significantly associated with the cnf1 virulence gene, but no significant relationship was observed with other virulence genes. The results of this study showed that these virulence genes in E. coli strains were not alone in determining the biofilm formation ability, but other factors such as environmental and genetic factors may affect biofilm formation (50). Similar studies were conducted by Naves et al., Tabasi et al., and Katongole et al. (16, 49, 51).
In this study, based on the method proposed by Clermont et al. (19) in 2000, we investigated the phylogenetic analysis of E. coli strains. Using Triplex PCR, E. coli strains can be divided into four groups: (1) B2, (2) D, (3) A, and (4) B1. Based on the results of this study, 73.6, 15.9, 5.5, and 4.86% were in groups B2, D, B1, and A, respectively. Uropathogenic E. coli strains mostly belonged to group B2 and to a lesser extent to group D. In this study, the distribution of virulence genes among the phylogenetic groups of E. coli was investigated. The prevalence of virulence genes belonging to group B2 was higher than that of other phylogenetic groups. The highest frequency of pathogenic genes in group B2 was related to fimH (94%), iutA (92%), ompT (90%), traT (89%), PAI (76%), aer (69%), iha (62%), cnf1 (41%), hlyA (34%), and iroN (31%). A significant correlation was observed between E. coli phylogenetic groups and aer (P = 0.019), iroN (P = 0.042), and ompT (P = 0.032) virulence genes (Table 3). The findings of our study were consistent with other studies. The high prevalence of fimbriae genes, iron acquisition receptors, and toxins in group B2 indicates the high pathogenic potential of these isolates as extraintestinal isolates (52-55).
A limitation of this study is that some of the virulence genes studied were involved in the formation of bacterial biofilms (including gelatinase formation and hemagglutination), and the selected community for the study was from a particular geographic region.
5.1. Conclusions
The results of this study showed that the frequency of virulence genes in the biofilm-forming strains was high. Also, strains that showed biofilm formation ability had higher resistance to antibiotics than strains without biofilm formation ability. Given the clinical importance of these virulence factors in the development and progression of UTIs and the role of biofilm formation in increasing bacterial resistance to antibiotics, the results of this study may help in the management of urinary tract infections and better medical interventions in this geographical area.
References
-
1.
Schifano E, Marazzato M, Ammendolia MG, Zanni E, Ricci M, Comanducci A, et al. Virulence behavior of uropathogenic Escherichia coli strains in the host model Caenorhabditis elegans. Microbiologyopen. 2019;8(6). e00756. [PubMed ID: 30381890]. [PubMed Central ID: PMC6562141]. https://doi.org/10.1002/mbo3.756.
-
2.
Usein CR, Damian M, Tatu-Chitoiu D, Capusa C, Fagaras R, Tudorache D, et al. Prevalence of virulence genes in Escherichia coli strains isolated from Romanian adult urinary tract infection cases. J Cell Mol Med. 2001;5(3):303-10. [PubMed ID: 12067489]. [PubMed Central ID: PMC6741423]. https://doi.org/10.1111/j.1582-4934.2001.tb00164.x.
-
3.
Boroumand M, Naghmachi M, Ghatee MA. Detection of phylogenetic groups and drug resistance genes of Escherichia coli causing urinary tract infection in Southwest Iran. Jundishapur J Microbiol. 2021;14(2). e112547. https://doi.org/10.5812/jjm.112547.
-
4.
Dong G, Li J, Chen L, Bi W, Zhang X, Liu H, et al. Effects of sub-minimum inhibitory concentrations of ciprofloxacin on biofilm formation and virulence factors of Escherichia coli. Braz J Infect Dis. 2019;23(1):15-21. [PubMed ID: 30796889]. https://doi.org/10.1016/j.bjid.2019.01.004.
-
5.
Tajbakhsh E, Ahmadi P, Abedpour-Dehkordi E, Arbab-Soleimani N, Khamesipour F. Biofilm formation, antimicrobial susceptibility, serogroups and virulence genes of uropathogenic E. coli isolated from clinical samples in Iran. Antimicrob Resist Infect Control. 2016;5:11. [PubMed ID: 27042294]. [PubMed Central ID: PMC4818419]. https://doi.org/10.1186/s13756-016-0109-4.
-
6.
Spencer JD, Schwaderer AL, Becknell B, Watson J, Hains DS. The innate immune response during urinary tract infection and pyelonephritis. Pediatr Nephrol. 2014;29(7):1139-49. [PubMed ID: 23732397]. [PubMed Central ID: PMC3800267]. https://doi.org/10.1007/s00467-013-2513-9.
-
7.
Donlan RM. Biofilms: microbial life on surfaces. Emerg Infect Dis. 2002;8(9):881-90. [PubMed ID: 12194761]. [PubMed Central ID: PMC2732559]. https://doi.org/10.3201/eid0809.020063.
-
8.
Sarowska J, Futoma-Koloch B, Jama-Kmiecik A, Frej-Madrzak M, Ksiazczyk M, Bugla-Ploskonska G, et al. Virulence factors, prevalence and potential transmission of extraintestinal pathogenic Escherichia coli isolated from different sources: Recent reports. Gut Pathog. 2019;11:10. [PubMed ID: 30828388]. [PubMed Central ID: PMC6383261]. https://doi.org/10.1186/s13099-019-0290-0.
-
9.
Boroumand M, Sharifi A, Manzouri L, Khoramrooz SS, Khosravani SA. Evaluation of pap and sfa genes relative frequency P and S fimbriae encoding of uropathogenic Escherichia coli isolated from hospitals and medical laboratories; Yasuj City, Southwest Iran. Iran Red Crescent Med J. 2019;21(8). https://doi.org/10.5812/ircmj.89499.
-
10.
Nikzad M, Mirnejad R, Babapour E. Evaluation of antibiotic resistance and biofilm formation ability uropathogenic E. coli (UPEC) isolated from pregnant women in Karaj. Iran J Med Microbiol. 2021;15(2):195-211. https://doi.org/10.30699/ijmm.15.2.195.
-
11.
Tarchouna M, Ferjani A, Ben-Selma W, Boukadida J. Distribution of uropathogenic virulence genes in Escherichia coli isolated from patients with urinary tract infection. Int J Infect Dis. 2013;17(6):e450-3. [PubMed ID: 23510539]. https://doi.org/10.1016/j.ijid.2013.01.025.
-
12.
Rashki A, Abdi HA, Shookohi M. Prevalence of genes encoding outer membrane virulence factors among fecal Escherichia coli isolates. Int J Basic Sci Med. 2017;2(1):52-7. https://doi.org/10.15171/ijbsm.2017.11.
-
13.
Neamati F, Firoozeh F, Saffari M, Zibaei M. Virulence genes and antimicrobial resistance pattern in uropathogenic Escherichia coli isolated from hospitalized patients in Kashan, Iran. Jundishapur J Microbiol. 2015;8(2). e17514. [PubMed ID: 25825647]. [PubMed Central ID: PMC4376973]. https://doi.org/10.5812/jjm.17514.
-
14.
Momtaz H, Karimian A, Madani M, Safarpoor Dehkordi F, Ranjbar R, Sarshar M, et al. Uropathogenic Escherichia coli in Iran: serogroup distributions, virulence factors and antimicrobial resistance properties. Ann Clin Microbiol Antimicrob. 2013;12:8. [PubMed ID: 23627669]. [PubMed Central ID: PMC3651382]. https://doi.org/10.1186/1476-0711-12-8.
-
15.
Behzadi P, Urban E, Gajdacs M. Association between biofilm-production and antibiotic resistance in uropathogenic Escherichia coli (UPEC): an in vitro study. Diseases. 2020;8(2). [PubMed ID: 32517335]. [PubMed Central ID: PMC7348726]. https://doi.org/10.3390/diseases8020017.
-
16.
Katongole P, Nalubega F, Florence NC, Asiimwe B, Andia I. Biofilm formation, antimicrobial susceptibility and virulence genes of Uropathogenic Escherichia coli isolated from clinical isolates in Uganda. BMC Infect Dis. 2020;20(1):453. [PubMed ID: 32600258]. [PubMed Central ID: PMC7325280]. https://doi.org/10.1186/s12879-020-05186-1.
-
17.
Freeman DJ, Falkiner FR, Keane CT. New method for detecting slime production by coagulase negative staphylococci. J Clin Pathol. 1989;42(8):872-4. [PubMed ID: 2475530]. [PubMed Central ID: PMC1142068]. https://doi.org/10.1136/jcp.42.8.872.
-
18.
Subramanian P, Shanmugam N, Sivaraman U, Kumar S, Selvaraj S. Antiobiotic resistance pattern of biofilm-forming uropathogens isolated from catheterised patients in Pondicherry, India. Australas Med J. 2012;5(7):344-8. [PubMed ID: 22905060]. [PubMed Central ID: PMC3412999]. https://doi.org/10.4066/AMJ.2012.1193.
-
19.
Clermont O, Bonacorsi S, Bingen E. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl Environ Microbiol. 2000;66(10):4555-8. [PubMed ID: 11010916]. [PubMed Central ID: PMC92342]. https://doi.org/10.1128/AEM.66.10.4555-4558.2000.
-
20.
Rahdar M, Rashki A, Miri HR, Rashki Ghalehnoo M. Detection of pap, sfa, afa, foc, and fim adhesin-encoding operons in uropathogenic Escherichia coli isolates collected from patients with urinary tract infection. Jundishapur J Microbiol. 2015;8(8). e22647. [PubMed ID: 26464770]. [PubMed Central ID: PMC4600570]. https://doi.org/10.5812/jjm.22647.
-
21.
Lee JH, Subhadra B, Son YJ, Kim DH, Park HS, Kim JM, et al. Phylogenetic group distributions, virulence factors and antimicrobial resistance properties of uropathogenic Escherichia coli strains isolated from patients with urinary tract infections in South Korea. Lett Appl Microbiol. 2016;62(1):84-90. [PubMed ID: 26518617]. https://doi.org/10.1111/lam.12517.
-
22.
Abdi HA, Rashki Ghalehnoo M. Virulence genes, genetic diversity, antimicrobial susceptibility and phylogenetic background of Escherichia coli isolates. Int J Enteric Pathog. 2015;3(3). https://doi.org/10.17795/ijep.25692.
-
23.
Cyoia PS, Rodrigues GR, Nishio EK, Medeiros LP, Koga VL, Pereira AP, et al. Presence of virulence genes and pathogenicity islands in extraintestinal pathogenic Escherichia coli isolates from Brazil. J Infect Dev Ctries. 2015;9(10):1068-75. [PubMed ID: 26517481]. https://doi.org/10.3855/jidc.6683.
-
24.
Oliveira FA, Paludo KS, Arend LN, Farah SM, Pedrosa FO, Souza EM, et al. Virulence characteristics and antimicrobial susceptibility of uropathogenic Escherichia coli strains. Genet Mol Res. 2011;10(4):4114-25. [PubMed ID: 22057993]. https://doi.org/10.4238/2011.October.31.5.
-
25.
Aazam K, Hassan M, Mahbobe M. Detection of uropathogenic Escherichia coli virulence factors in patients with urinary tract infections in Iran. Afr J Microbiol Res. 2012;6(39):6811-6. https://doi.org/10.5897/ajmr12.1462.
-
26.
Kot B, Wicha J, Grużewska A, Piechota M, Wolska K, Obrębska M. Virulence factors, biofilm-forming ability, and antimicrobial resistance of urinary Escherichia coli strains isolated from hospitalized patients. Turk J Med Sci. 2016;46(6):1908-14. [PubMed ID: 28081347]. https://doi.org/10.3906/sag-1508-105.
-
27.
Ejrnaes K. Bacterial characteristics of importance for recurrent urinary tract infections caused by Escherichia coli. Dan Med Bull. 2011;58(4):B4187. [PubMed ID: 21466767].
-
28.
Hall CW, Mah TF. Molecular mechanisms of biofilm-based antibiotic resistance and tolerance in pathogenic bacteria. FEMS Microbiol Rev. 2017;41(3):276-301. [PubMed ID: 28369412]. https://doi.org/10.1093/femsre/fux010.
-
29.
Mittal S, Sharma M, Chaudhary U. Biofilm and multidrug resistance in uropathogenic Escherichia coli. Pathog Glob Health. 2015;109(1):26-9. [PubMed ID: 25605466]. [PubMed Central ID: PMC4445292]. https://doi.org/10.1179/2047773215Y.0000000001.
-
30.
Karam MRA, Habibi M, Bouzari S. Relationships between virulence factors and antimicrobial resistance among Escherichia coli isolated from urinary tract infections and commensal isolates in Tehran, Iran. Osong Public Health Res Perspect. 2018;9(5):217-24. [PubMed ID: 30402376]. [PubMed Central ID: PMC6202021]. https://doi.org/10.24171/j.phrp.2018.9.5.02.
-
31.
Poursina F, Sepehrpour S, Mobasherizadeh S. Biofilm Formation in Nonmultidrug-resistant Escherichia coli Isolated from Patients with Urinary Tract Infection in Isfahan, Iran. Adv Biomed Res. 2018;7:40. [PubMed ID: 29657925]. [PubMed Central ID: PMC5887692]. https://doi.org/10.4103/abr.abr_116_17.
-
32.
Sudheendra K, Basavaraj P. Analysis of antibiotic sensitivity profile of biofilm-forming uropathogenic Escherichia coli. J Nat Sci Biol Med. 2018;9(2):175.
-
33.
Neupane S, Pant ND, Khatiwada S, Chaudhary R, Banjara MR. Correlation between biofilm formation and resistance toward different commonly used antibiotics along with extended spectrum beta lactamase production in uropathogenic Escherichia coli isolated from the patients suspected of urinary tract infections visiting Shree Birendra Hospital, Chhauni, Kathmandu, Nepal. Antimicrob Resist Infect Control. 2016;5:5. [PubMed ID: 26885364]. [PubMed Central ID: PMC4754952]. https://doi.org/10.1186/s13756-016-0104-9.
-
34.
Poovendran P, Ramanathan N. In vitro study on antibiotic susceptibility pattern of biofilm producing uropathogenic Escherichia coli isolates and their molecular characterization. Asian J Pharm Clin Res. 2014;7(3):181-5.
-
35.
Niveditha S, Pramodhini S, Umadevi S, Kumar S, Stephen S. The isolation and the biofilm formation of uropathogens in the patients with catheter associated urinary tract infections (UTIs). J Clin Diagn Res. 2012;6(9):1478-82. [PubMed ID: 23285434]. [PubMed Central ID: PMC3527774]. https://doi.org/10.7860/JCDR/2012/4367.2537.
-
36.
Ponnusamy P, Natarajan V, Sevanan M. In vitro biofilm formation by uropathogenic Escherichia coli and their antimicrobial susceptibility pattern. Asian Pac J Trop Med. 2012;5(3):210-3. [PubMed ID: 22305786]. https://doi.org/10.1016/S1995-7645(12)60026-1.
-
37.
Schiebel J, Bohm A, Nitschke J, Burdukiewicz M, Weinreich J, Ali A, et al. Genotypic and phenotypic characteristics associated with biofilm formation by human clinical Escherichia coli isolates of different pathotypes. Appl Environ Microbiol. 2017;83(24). [PubMed ID: 28986371]. [PubMed Central ID: PMC5717203]. https://doi.org/10.1128/AEM.01660-17.
-
38.
Fattahi S, Kafil HS, Nahai MR, Asgharzadeh M, Nori R, Aghazadeh M. Relationship of biofilm formation and different virulence genes in uropathogenic Escherichia coli isolates from Northwest Iran. GMS Hyg Infect Control. 2015;10:Doc11. [PubMed ID: 26213679]. [PubMed Central ID: PMC4512245]. https://doi.org/10.3205/dgkh000254.
-
39.
Awoke N, Kassa T, Teshager L. Magnitude of biofilm formation and antimicrobial resistance pattern of bacteria isolated from urinary catheterized inpatients of Jimma university medical center, southwest Ethiopia. Int J Microbiol. 2019;2019:5729568. [PubMed ID: 30881456]. [PubMed Central ID: PMC6387724]. https://doi.org/10.1155/2019/5729568.
-
40.
Baur D, Gladstone BP, Burkert F, Carrara E, Foschi F, Dobele S, et al. Effect of antibiotic stewardship on the incidence of infection and colonisation with antibiotic-resistant bacteria and Clostridium difficile infection: a systematic review and meta-analysis. Lancet Infect Dis. 2017;17(9):990-1001. [PubMed ID: 28629876]. https://doi.org/10.1016/S1473-3099(17)30325-0.
-
41.
Mukonzo JK, Namuwenge PM, Okure G, Mwesige B, Namusisi OK, Mukanga D. Over-the-counter suboptimal dispensing of antibiotics in Uganda. J Multidiscip Healthc. 2013;6:303-10. [PubMed ID: 23990728]. [PubMed Central ID: PMC3753154]. https://doi.org/10.2147/JMDH.S49075.
-
42.
Amadu D, Nwabuisi C, Yunusa T, Nasir I, Oladejo JM, Seibu E, et al. Prevalence and associated factors associated with Uropathogenic Escherichia coli isolates from catheterized persons at Ilorin Tertiary Hospital, Nigeria. Afro-Egyptian Journal of Infectious and Endemic Diseases. 2019;9(2):119-28. https://doi.org/10.21608/aeji.2019.11367.1019.
-
43.
Shokouhi Mostafavi SK, Najar-Peerayeh S, Mohabbati Mobarez A, Kardoust Parizi M. Serogroup distribution, diversity of exotoxin gene profiles, and phylogenetic grouping of CTX-M-1- producing uropathogenic Escherichia coli. Comp Immunol Microbiol Infect Dis. 2019;65:148-53. [PubMed ID: 31300106]. https://doi.org/10.1016/j.cimid.2019.05.003.
-
44.
Robino L, Scavone P, Araujo L, Algorta G, Zunino P, Pirez MC, et al. Intracellular bacteria in the pathogenesis of Escherichia coli urinary tract infection in children. Clin Infect Dis. 2014;59(11):e158-64. [PubMed ID: 25091303]. [PubMed Central ID: PMC4650771]. https://doi.org/10.1093/cid/ciu634.
-
45.
Navidinia M, Najar Peerayeh S, Fallah F, Bakhshi B. Phylogenetic groups and pathogenicity island markers in Escherichia coli isolated from children. Jundishapur J Microbiol. 2013;6(10). e8362. https://doi.org/10.5812/jjm.8362.
-
46.
Er DK, Dundar D, Uzuner H, Osmani A. Relationship between phylogenetic groups, antibiotic resistance and patient characteristics in terms of adhesin genes in cystitis and pyelonephritis isolates of Escherichia coli. Microb Pathog. 2015;89:188-94. [PubMed ID: 26518125]. https://doi.org/10.1016/j.micpath.2015.10.014.
-
47.
Horcajada JP, Soto S, Gajewski A, Smithson A, Jimenez de Anta MT, Mensa J, et al. Quinolone-resistant uropathogenic Escherichia coli strains from phylogenetic group B2 have fewer virulence factors than their susceptible counterparts. J Clin Microbiol. 2005;43(6):2962-4. [PubMed ID: 15956432]. [PubMed Central ID: PMC1151912]. https://doi.org/10.1128/JCM.43.6.2962-2964.2005.
-
48.
Farajzadah Sheikh A, Goodarzi H, Yadyad MJ, Aslani S, Amin M, Jomehzadeh N, et al. Virulence-associated genes and drug susceptibility patterns of uropathogenic Escherichia coli isolated from patients with urinary tract infection. Infect Drug Resist. 2019;12:2039-47. [PubMed ID: 31410031]. [PubMed Central ID: PMC6646852]. https://doi.org/10.2147/IDR.S199764.
-
49.
Naves P, del Prado G, Huelves L, Gracia M, Ruiz V, Blanco J, et al. Correlation between virulence factors and in vitro biofilm formation by Escherichia coli strains. Microb Pathog. 2008;45(2):86-91. [PubMed ID: 18486439]. https://doi.org/10.1016/j.micpath.2008.03.003.
-
50.
Reisner A, Krogfelt KA, Klein BM, Zechner EL, Molin S. In vitro biofilm formation of commensal and pathogenic Escherichia coli strains: impact of environmental and genetic factors. J Bacteriol. 2006;188(10):3572-81. [PubMed ID: 16672611]. [PubMed Central ID: PMC1482849]. https://doi.org/10.1128/JB.188.10.3572-3581.2006.
-
51.
Tabasi M, Asadi Karam MR, Habibi M, Yekaninejad MS, Bouzari S. Phenotypic assays to determine virulence factors of uropathogenic Escherichia coli (UPEC) isolates and their correlation with antibiotic resistance pattern. Osong Public Health Res Perspect. 2015;6(4):261-8. [PubMed ID: 26473094]. [PubMed Central ID: PMC4588432]. https://doi.org/10.1016/j.phrp.2015.08.002.
-
52.
Alizade H, Ghanbarpour R, Aflatoonian MR, Abdollahi H. Determination of phylogenetic background, fimbrial genes, and antibiotic susceptibility of Escherichia coli isolates from urinary tract infections in Bam region, Iran. Comp Clin Path. 2013;23(5):1253-7. https://doi.org/10.1007/s00580-013-1771-z.
-
53.
Derakhshandeh A, Firouzi R, Motamedifar M, Motamedi Boroojeni A, Bahadori M, Arabshahi S, et al. Distribution of virulence genes and multiple drug-resistant patterns amongst different phylogenetic groups of uropathogenic Escherichia coli isolated from patients with urinary tract infection. Lett Appl Microbiol. 2015;60(2):148-54. [PubMed ID: 25355175]. https://doi.org/10.1111/lam.12349.
-
54.
Cristea VC, Gheorghe I, Czobor Barbu I, Popa LI, Ispas B, Grigore GA, et al. Snapshot of phylogenetic groups, virulence, and resistance markers in Escherichia coli uropathogenic strains isolated from outpatients with urinary tract infections in Bucharest, Romania. Biomed Res Int. 2019;2019:5712371. [PubMed ID: 31236408]. [PubMed Central ID: PMC6545812]. https://doi.org/10.1155/2019/5712371.
-
55.
Hyun M, Lee JY, Kim HA. Differences of virulence factors, and antimicrobial susceptibility according to phylogenetic group in uropathogenic Escherichia coli strains isolated from Korean patients. Research Square. 2021;Preprint. https://doi.org/10.21203/rs.3.rs-575640/v1.