Abstract
Background:
The emergence of antibiotic-resistant Staphylococcus aureus strains is one of the major concerns about the various staphylococcal infections. Vancomycin is one the most important effective antibiotics on staphylococcal lethal infections. To date, vancomycin-resistant strains are increasingly isolated in different parts of the world, and it is alerting.Objectives:
The current study was designed to evaluate the prevalence, and antibiotic susceptibility pattern of methicillin-resistant S. aureus (MRSA) and vancomycin-resistant S. aureus (VRSA) isolates in the main tertiary hospital of Bojnurd, Iran.Methods:
Staphylococcus aureus isolates were collected from different clinical samples in Imam Reza Hospital of Bojnurd. After identification of isolates through using conventional methods, they were evaluated by agar screening, disk diffusion, and minimum inhibitory concentration (MIC) methods to determine resistance to vancomycin and methicillin. We also performed polymerase chain reaction (PCR) for the detection of mecA, mecC, vanA, and vanB genes. After confirmation of vancomycin resistance, genetic analysis was performed using SCCmec, agr, and spa typing, and multilocus sequence typing (MLST) methods on VRSA isolates.Results:
We found four vancomycin-resistant isolates (1.29%). Also, 75% of isolates were resistant to cefoxitin. Using the PCR method, mecA was found in 73%, mecC in 0.64%, and vanA in 1.29% of isolates. Interestingly, we found two mecC positive isolates in MRSA isolates. The alpha-hemolysin (81.81%) and enterotoxin C (27%) had the highest and lowest toxins percentage, respectively. Among mecA positive isolates, SCCmec IV (37%), SCCmec III (31.27%), SCCmec I (14%), SCCmec II (11%), and SCCmec V (5.7%) were the most prevalent SCCmec types, respectively. It should be noted that the two mecC positive isolates belonged to SCCmec XI. Agr I (76.29%) was the highest agr type. We recognized t037 as the dominant spa type, and ST239, ST6, ST97, and ST8 were found in VRSA isolates.Conclusions:
In our study, the frequency of mecA genes in MRSA isolates was very high. It seems that the resistant isolates belonged to endemic clones of Iran.Keywords
Staphylococcus aureus Multilocus Sequence Typing Spa Typing mecC Vancomycin
1. Background
Staphylococcus aureus is one of the most important opportunistic human pathogens. It’s a causative agent of a wide range of infections, from skin infections to life-threatening systemic infections. This bacterium can also lead to animal infections that can transfer to a human directly or by animal products such as meat, milk, etc. (1). Its pathogenicity is related to toxins and different enzymes such as staphylococcal enterotoxins, exfoliative toxins (ETs), and toxic shock syndrome toxin-1 (TSST-1) genes (2-5). The emergence of staphylococci resistant to antimicrobial agents is an issue of world concern the main cause of which may be an arbitrary use of broad-spectrum and unawareness of them (6).
In most cases, antimicrobial resistance is achieved due to the resistance gene transfer between bacteria from the same or different strains. For example, the methicillin resistance is achieved by the transfer of the mecA gene by a mobile genetic element staphylococcal cassette chromosome (SCC), and vancomycin resistance is achieved by vanA gene transfer using Tn1545 transposon. This type of resistance is occasional, but it is becoming a public health challenge due to its ability of fast dissemination (7). Recently, some European researchers found mecA- methicillin-resistant S. aureus (MRSA) isolates. They found analogs named mecC. The origins of mecC are not clear yet, but there is evidence that it can be transmitted through contact with animals (8). Most of the patients with mecC MRSA lived in rural areas (9).
2. Objectives
Regarding the importance of S. aureus related community and hospital-acquired infections, the current study was designed to check the prevalence and antibiotic susceptibility pattern of MRSA and vancomycin-resistant S. aureus (VRSA) isolates in the main tertiary hospital of Bojnurd, Iran between 2013 and 2018.
3. Methods
3.1. Sample Collection and Identification
Samples were isolated from clinical specimens, including blood, tissue, urine, sputum, wound, peritoen, nasal, trachea, bronchi, abscess, cerebrospinal fluid (CSF), catheter, throat, eye, and synovial. The confirmatory tests identified 308 clinical specimens as S. aureus isolates in Imam Reza hospital of Bojnurd. Staphylococcus aureus isolates were identified by standard biochemical and microbiological tests including Gram stain tests, catalase, mannitol fermentation, DNase, and slide and tube coagulase tests (10).
3.2. Antimicrobial Susceptibility Tests
Antibiotic susceptibility test was carried out on all isolates using Kirby-Bauer method based on 2018 guidelines by the Clinical and Laboratory Standards Institute (CLSI) (11). The antimicrobial disks used were vancomycin 5 µg (V), cefoxitin 30 µg (FOX), ciprofloxacin 5 µg (CIP), tetracycline 30 µg (TE), co-trimoxazole 1.25 µg (SXT), gentamicin 10 µg (GM), clindamycin 2 µg (CC), cefotaxime 30 µg (CEF), and rifampin 5 µg (R) (MAST DISKSTM, UK).
3.3. Agar Screen Test
All isolates were screened for oxacillin and vancomycin resistance using the agar screening method. Methicillin resistance was defined as the capability of growth in agar screening media including 4% NaCl supplemented with 6 µg/mL oxacillin, whereas vancomycin resistance was defined as the capability of growth in agar screening media including 6 µg/mL vancomycin (12).
3.4. Determination of Minimum Inhibitory Concentration
Resistant isolates were determined by measuring the minimum inhibitory concentration (MIC) of oxacillin and vancomycin. Oxacillin resistance was defined as a MIC ≥ 4 µg/mL, and vancomycin resistance was defined as MIC ≥ 16 µg/mL (12).
3.5. Genomic DNA Extraction
Genomic DNAs of all 308 S. aureus isolates were extracted using a QiaAmp DNA Mini Kit (QIAGEN, Germany). According to the manufacturer’s protocol for bacterial cells, we added lysostaphin at a final concentration of 30 µg/mL in the lysis buffer (13).
3.6. Detection of Resistance Genes
The presence of the mecA, mecC, vanA, and vanB genes was evaluated using the below primers as previously described (Table 1) (12-15).
Primer Sequence Used in This Study
| Gene | Primer Sequence | Product Size (bp) |
|---|---|---|
| mecA | 5'- AGAAGATGGTATGTGGAAGTTAG-3' | 584 |
| 5'- ATGTATGTGCGATTGTATTGC-3' | ||
| vanA | 5'- GGCAAGTCAGGTGAAGATG-3' | 713 |
| 5'- ATCAAGCGGTCAATCAGTTC-3' | ||
| vanB | 5'-ACGGAATGGGAAGCCGA-3' | 647 |
| 5'-TGCACCCGATTTCGTTC-3' | ||
| mecC | 5'-CATTAAAATCAGAGCGAGGC-3' | 188 |
| 5'-TGGCTGAACCCATTTTTGAT-3' | ||
| Sec | 5'-GGGAATGTTGGATGAAGG-3' | 900 |
| 5'-AGGCAAGCACCGAAGTAC-3' | ||
| tst1 | 5'-TTATCGTAAGCCCTTTGTTG-3' | 398 |
| 5'-TAAAGGTAGTTCTATTGGAGTAGG-3' | ||
| pvl | 5'-GGAAACATTTATTCTGGCTATAC-3' | 502 |
| 5'-CTGGATTGAAGTTACCTCTGG-3' | ||
| hla | 5'-CGGTACTACAGATATTGGAAGC-3' | 744 |
| 5'-TGGTAATCATCACGAACTCG-3' |
3.7. Multiplex Polymerase Chain Reaction for Detection of Toxin Genes
The specific primers for detection of the S. aureus enterotoxin C (sec), toxic shock syndrome toxin 1 (TSST-1), Panton-Valentine leukocidin (pvl), and alpha-hemolysin (hla) are listed in Table 1 (12) All S. aureus isolates were evaluated for these toxin genes.
3.8. Multiplex PCR for Agr and SCCmec Typing
The agr and SCCmec typing were performed on all 308 S. aureus isolates using Multiplex PCR as previously described (12, 16).
3.9. Multilocus Sequence Typing
Multilocus sequence typing (MLST) was performed on four VRSA and two mecC positive isolates based on amplification and sequencing of internal fragments of housekeeping genes S. aureus (arc, aro, glp, gmk, pta, tpi, and yqi genes) as previously described (12).
3.10. Spa Typing
Spa types of four VRSA and two mecC positive isolates were determined by PCR and sequencing of the polymorphic X region of spa gene as previously described (http://spaserver.ridom.de/) (12, 17). We also used the spa gene as an internal control gene for S. aureus verification in mecC and vanA harboring isolates.
4. Results
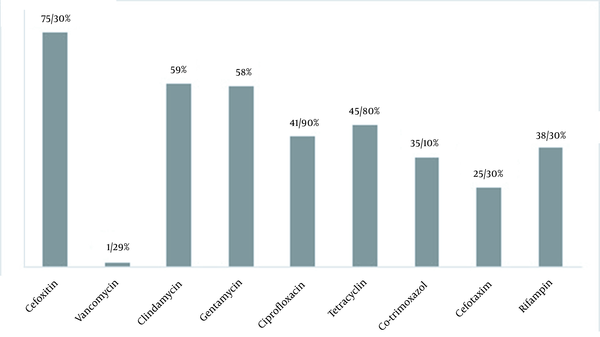
Using confirmatory tests, 308 clinical specimens were identified as S. aureus isolates. The results in antibiogram analysis showed that S. aureus resistance to cefoxitin (75.3%) had the highest resistance rate, followed by clindamycin (59%) and gentamicin (58%). The lowest resistance was seen for vancomycin (1.29%) (Figure 1). We found four vanA harboring vancomycin-resistant isolates with vancomycin MIC ≥ 16 μg/mL. The vanB gene PCR was negative in all these VRSA isolates. Interestingly, in cefoxitin isolates, we found two mecA-/mecC+ cases. The genetic characteristics of VRSA and mecC+ isolates are listed in Tables 2 and 3, respectively.
TSST was the most prevalent toxin in VRSA isolates, and hla was found in both mecC+ isolates. Hla was found in 81.8%, TSST in 33.8%, pvl in 29.2%, and sec in 27.3% of all 308 isolates. AgrI (76.29%) was the most prevalent agr type. Of four VRSA isolates, two were also MRSA and belonged to SCCmecI and III, and mecC+ isolates belonged to SCCmecXI. Moreover, in all MRSA isolates the most prevalent SCCmec types were SCCmecIV (37%), SCCmecIII (31.27%), SCCmec I (14%), SCCmecII (11%), SCCmecV (5.7%), and SCCmecXI, respectively. More details regarding VRSA and mecC+ isolates are presented in Tables 2 and 3.
Antimicrobial resistance of Staphylococcus aureus isolates (n = 308)
Genetic and Phenotypic Characteristics of vancomycin-resistant Staphylococcus aureus Isolates
| Source | Resistance | Vancomycin MIC | Resistance Gene | SCCmec Type | Agr | Toxin Gene | Spa Type | ST | |
|---|---|---|---|---|---|---|---|---|---|
| VRSA1 | Blood | Cef, gen, tet | 32 | mecA, vanA | III | I | Pvl, TSST, etc, hla | t037 | ST239 |
| VRSA2 | Blood | Rif, cli | 16 | vanA | - | I | TSST | t189 | ST239 |
| VRSA3 | trachea | Cip, cli, cot, tet | 16 | vanA | - | I | TSST | t304 | ST6 |
| VRSA4 | wound | Cef, cli, cot | 32 | mecA, vanA | I | NT | - | t7688 | ST97 |
Genetic and Phenotypic Characteristics of mecC+ Isolates
| Resistance | Oxacillin MIC | Resistance Gene | SCCmec Type | Agr | Toxin Gene | Spa Type | ST | |
|---|---|---|---|---|---|---|---|---|
| mecC1 | cli, cef | 64 | mecC | XI | I | hla | t230 | ST8 |
| mecC2 | cef, cli, cot | 32 | mecC | XI | I | hla | t037 | ST239 |
5. Discussion
Due to the importance of Staphylococcal infections and their increasing antimicrobial resistance and also the undeniable role in human hospital and community-acquired infections, in this study, we evaluated the antimicrobial resistance of S. aureus isolates in various clinical samples in Bojnurd, Iran, from 2013 to 2018. The prevalence of VRSA has increased in recent years in different parts of the world. Its total prevalence was 2% before 2006, 5% in 2006 - 2014, and 7% in 2015 - 2020. The prevalence of VRSA was 5% in Asia, 1% in Europe, 4% in America, 3% in South America, and 16% in Africa (18). Interestingly, we found four VRSA isolates in samples received in different years. These isolates were resistant to cefotaxime, gentamycin, tetracycline, rifampin, clindamycin, ciprofloxacin, and co-trimoxazole (Table 2).
Two isolates were MRSA, and two other isolates were not harboring the mecA gene. The MIC of vancomycin was 16 - 32 µg/mL, although in previously found VRSA isolates in northeastern Iran, we witnessed a higher MIC for this antibiotic (12). The vancomycin MIC in mecA+ VRSA is twofold (32 µg/mL) as mecA- VRSA was 16 µg/mL. This may be due to the effect of mecA gene product on cell wall precursors (depsipeptides) that are the substrate of vanA gene product. The low-affinity PBP2A cannot utilize the depsipeptides, while in mecA negative strains PBP2 can utilize the cell wall precursors including depsipeptides (19). This isolate belonged to ST239 and spa t037, whereas our isolates belonged to other different ST/spa types in addition to ST239/t037. We have ST239/t189, ST6/t304, and ST97/t7688 isolates. It should be noted that these strains were isolated at different times, and maybe this variety indicates that the strains of our region have acquired resistance genes separately and do not have a common origin.
As discussed in many previously published articles, the source of vanA gene can be vancomycin-resistant enterococci or other vancomycin-resistant staphylococci (20). If the VRSA isolates are the source of resistance genes, we should find the same clones in different samples; but regarding the diversity of genetic clones of our isolates, we may Predict the independent acquisition of resistance in different isolates. This is an alarming issue, as it may indicate a much higher prevalence and potential for further changes in these strains in the region. It should also be noted that Bojnurd is bordered by Turkmenistan, and it is also on the way to the city of Mashhad, which is a holy city visited by many tourists from all Islamic countries. This specific geographical position can lead to the entrance of different clones of bacteria to this area.
ST239/t037 is the endemic Asian clone previously found in different regions of Iran, and other clones such as ST97/t7688 are also found in this region. The acquisition of the vanA gene in endemic clones may be due to the co-infection of these clones with vancomycin-resistant enterococcus spp. as a vanA gene cluster source or other vancomycin-resistant staphylococci spp. Another noteworthy point is the source of strain isolation.
Two VRSA samples were isolated from the blood samples; one of them was isolated from the trachea, and the other one was isolated from a burn wound. This diversity of sources of sample isolation can indicate their high ability to spread and cause infection in different parts of the body, from superficial wound infections to systemic infections such as bloodstream infections. We previously found VRSA in the trachea sample of a dead patient in Mashhad, northeastern Iran (12). The resistance to oxacillin in wound isolated S. aureus strians was reported in many reports worldwide (21-23), but resistance to vancomycin in these isolates is not prevalent (24). In recent years, some researchers reported mecA- MRSA isolates. They found the mecA analogs gene (mecC) that acts like mecA. MecC is located within the SCCmec element, and it was found in some animal-related Staphylococci such as S. sciuri and S. xylosus (25, 26). This gene has 68.7% nucleotide homology with mecA and is located in a new SCCmec element called SCCmec XI. It has proved to be resistant to beta-lactams phenotypically (27, 28).
The mecC gene was reported in isolates from humans and dairy cattle, and some recent reports showed the prevalence of mecC mediated methicillin resistance among human MRSA isolates (29). Some studies reported that mecC harboring MRSA has a lower oxacillin MIC than their mecA harboring isolates (8), but interestingly in our isolates, MIC was 32 and 64 mg/mL while MIC of ≥ 4 μg/mL was found in about 99% of mecA+ isolates. The mecC+ isolates were mainly found in Europe and rarely reported in other countries.
Most previously found isolates belonged to animal-related clones, but interestingly our isolates belonged to human-related endemic Asian clones, including ST239 and ST8. Thus, it could be concluded that these clones were not transferred from European sources to Iran. In the first report in Iran, mecC+ MRSA isolates belonged to ST130 and ST599 and spa types t843 and t5930, while our isolates belonged to ST239/t037 and ST8/t230 clones. It should be noted that most people living in North Khorasan province reside in rural areas, and animal husbandry is one of the main occupations in this area. Close human-animal contact may have led to gene transfer between human-related strains and animal-related strains. However, this assumption needs further evaluations.
5.1. Conclusions
In this study, vancomycin resistance was observed in four strains belonging to endemic clones. It’s alarming because it may be a sign of genetic evolution and the spreading of these resistant isolates. We also found mecC+ MRSA isolates in human-related clones. We predict more distribution of these isolates and suggest to revise in molecular methods of MRSA detection in this area. More studies with larger sample size are required to confirm these results.
Acknowledgements
References
-
1.
Xing X, Zhang Y, Wu Q, Wang X, Ge W, Wu C. Prevalence and characterization of Staphylococcus aureus isolated from goat milk powder processing plants. Food Control. 2016;59:644-50. https://doi.org/10.1016/j.foodcont.2015.06.042.
-
2.
Akpaka PE, Monecke S, Swanston WH, Rao AC, Schulz R, Levett PN. Methicillin sensitive Staphylococcus aureus producing Panton-Valentine leukocidin toxin in Trinidad & Tobago: a case report. J Med Case Rep. 2011;5(5):1-5. [PubMed ID: 21507214]. [PubMed Central ID: PMC3110132]. https://doi.org/10.1186/1752-1947-5-157.
-
3.
Gordon RJ, Lowy FD. Pathogenesis of methicillin-resistant Staphylococcus aureus infection. Clin Infect Dis. 2008;46 Suppl 5:350-9. [PubMed ID: 18462090]. [PubMed Central ID: PMC2474459]. https://doi.org/10.1086/533591.
-
4.
Rahimi F, Bouzari M, Katouli M, Pourshafie MR. Antibiotic Resistance Pattern of Methicillin Resistant and Methicillin Sensitive Staphylococcus aureus Isolates in Tehran, Iran. Jundishapur J Microbiol. 2013;6(2):144-9. https://doi.org/10.5812/jjm.4896.
-
5.
Liu B. Prevalence, resistance pattern, and molecular characterization of Staphylococcus aureus isolates from healthy animals and sick populations in Henan Province, China. Gut pathogens. 2018;10(1):1-13.
-
6.
McGuinness WA, Malachowa N, DeLeo FR. Focus: infectious diseases: vancomycin resistance in Staphylococcus aureus. Yale J Biol Med. 2017;90(2):269.
-
7.
Hiramatsu K, Katayama Y, Matsuo M, Sasaki T, Morimoto Y, Sekiguchi A, et al. Multi-drug-resistant Staphylococcus aureus and future chemotherapy. J Infect Chemother. 2014;20(10):593-601. [PubMed ID: 25172776]. https://doi.org/10.1016/j.jiac.2014.08.001.
-
8.
Paterson GK, Harrison EM, Holmes MA. The emergence of mecC methicillin-resistant Staphylococcus aureus. Trends Microbiol. 2014;22(1):42-7. [PubMed ID: 24331435]. [PubMed Central ID: PMC3989053]. https://doi.org/10.1016/j.tim.2013.11.003.
-
9.
Lindgren AK, Gustafsson E, Petersson AC, Melander E. Methicillin-resistant Staphylococcus aureus with mecC: a description of 45 human cases in southern Sweden. Eur J Clin Microbiol Infect Dis. 2016;35(6):971-5. [PubMed ID: 27010813]. https://doi.org/10.1007/s10096-016-2624-x.
-
10.
Saadat S, Solhjoo K, Norooz-Nejad MJ, Kazemi A. VanA and VanB positive vancomycin-resistant Staphylococcus aureus among clinical isolates in Shiraz, South of Iran. Oman Med J. 2014;29(5):335. [PubMed ID: 25337309]. [PubMed Central ID: PMC4202220]. https://doi.org/10.5001/omj.2014.90.
-
11.
Wayne PA. Clinical and Laboratory Standards Institute: Performance standards for antimicrobial susceptibility testing: 20th informational supplement. CLSI document; 2018. p. 100-20.
-
12.
Azimian A, Havaei SA, Fazeli H, Naderi M, Ghazvini K, Samiee SM, et al. Genetic characterization of a vancomycin-resistant Staphylococcus aureus isolate from the respiratory tract of a patient in a university hospital in northeastern Iran. J Clin Microbiol. 2012;50(11):3581-5. [PubMed ID: 22933598]. [PubMed Central ID: PMC3486207]. https://doi.org/10.1128/JCM.01727-12.
-
13.
Havaei SA, Azimian A, Fazeli H, Naderi M, Ghazvini K, Samiee SM, et al. Genetic characterization of methicillin resistant and sensitive, vancomycin intermediate Staphylococcus aureus strains isolated from different Iranian hospitals. ISRN Microbiol. 2012;2012:215275. [PubMed ID: 23762750]. [PubMed Central ID: PMC3664199]. https://doi.org/10.5402/2012/215275.
-
14.
Havaei SA, Moghim S, Shahin M, Azimian A, Ghanbari F, Shokri D, et al. [A comparison between polymerase chain reaction, oxacillin agar dilution and cefoxitin disk diffusion methods in detection of methicillin resistance in staphylococcus aureus]. J Isfahan Med Sch. 2013;31(232):466-74. Persian.
-
15.
Azimian A, Asghar Havaei S, Fazeli H, Naderi M, Ghazvini K, Mirab Samiee S, et al. Genetic analysis of a vancomycin-resistant Staphylococcus aureus strain isolated in Iran. mBio. 2012;3(6). [PubMed ID: 23170001]. [PubMed Central ID: PMC3509435]. https://doi.org/10.1128/mBio.00442-12.
-
16.
Ito T, Kuwahara-Arai K, Katayama Y, Uehara Y, Han X, Kondo Y, et al. Staphylococcal Cassette Chromosome mec (SCCmec) analysis of MRSA. Springer; 2014. p. 131-48.
-
17.
Harmsen D, Claus H, Witte W, Rothganger J, Claus H, Turnwald D, et al. Typing of methicillin-resistant Staphylococcus aureus in a university hospital setting by using novel software for spa repeat determination and database management. J Clin Microbiol. 2003;41(12):5442-8. [PubMed ID: 14662923]. [PubMed Central ID: PMC309029]. https://doi.org/10.1128/JCM.41.12.5442-5448.2003.
-
18.
Wu Q, Sabokroo N, Wang Y, Hashemian M, Karamollahi S, Kouhsari E. Systematic review and meta-analysis of the epidemiology of vancomycin-resistance Staphylococcus aureus isolates. Antimicrob Resist Infect Control. 2021;10(1):1-13. [PubMed ID: 34193295]. [PubMed Central ID: PMC8247230]. https://doi.org/10.1186/s13756-021-00967-y.
-
19.
Severin A, Tabei K, Tenover F, Chung M, Clarke N, Tomasz A. High level oxacillin and vancomycin resistance and altered cell wall composition in Staphylococcus aureus carrying the staphylococcal mecA and the enterococcal vanA gene complex. J Biol Chem. 2004;279(5):3398-407. [PubMed ID: 14613936]. https://doi.org/10.1074/jbc.M309593200.
-
20.
Besharati R, Ghafouri M, Safamanesh S, Khosrojerdi M, Ghazvini K, Nojumi S, et al. Molecular epidemiology of Panton-valentine leukocidin harboring hospital-associated methicillin-resistant Staphylococcus aureus in septicemic children, Northeastern Iran, Bojnurd. Jundishapur J Microbiol. 2019;12(2). https://doi.org/10.5812/jjm.68183.
-
21.
Wankhade AB, Panda S, Hathiwala R, Keche Y. Study of antibiotic resistance profiling of Staphylococcus aureus isolated from clinical specimens of the patients attending a tertiary teaching hospital from Chhattisgarh. Int J Res Med Sci. 2017;5(11). https://doi.org/10.18203/2320-6012.ijrms20174924.
-
22.
Tambekar DH, Dhanorkar DV, Gulhane SR, Dudhane MN. Prevalence and antimicrobial susceptibility pattern of methicillin resistant Staphylococcus aureus from healthcare and community associated sources. Afr J Infect Dis. 2008;1(1):52-6. https://doi.org/10.4314/ajid.v1i1.42086.
-
23.
Joshi SG, Paff M, Friedman G, Fridman G, Fridman A, Brooks AD. Control of methicillin-resistant Staphylococcus aureus in planktonic form and biofilms: a biocidal efficacy study of nonthermal dielectric-barrier discharge plasma. Am J Infect Control. 2010;38(4):293-301. [PubMed ID: 20085853]. https://doi.org/10.1016/j.ajic.2009.11.002.
-
24.
Ghanbari F, Saberianpour S, Zarkesh-Esfahani F, Ghanbari N, Taraghian A, Khademi F. Staphylococcal cassette chromosome mec (SCCmec) typing of methicillin-resistant Staphylococcus aureus strains isolated from community- and hospital-acquired infections. Avicenna J Clin Microbiol Infect. 2017;4(2):42244. https://doi.org/10.5812/ajcmi.42244.
-
25.
Harrison EM, Paterson GK, Holden MT, Morgan FJ, Larsen AR, Petersen A, et al. A Staphylococcus xylosus isolate with a new mecC allotype. Antimicrob Agents Chemother. 2013;57(3):1524-8. [PubMed ID: 23274660]. [PubMed Central ID: PMC3591899]. https://doi.org/10.1128/AAC.01882-12.
-
26.
Harrison EM, Paterson GK, Holden MT, Ba X, Rolo J, Morgan FJ, et al. A novel hybrid SCCmec-mecC region in Staphylococcus sciuri. J Antimicrob Chemother. 2014;69(4):911-8. [PubMed ID: 24302651]. [PubMed Central ID: PMC3956370]. https://doi.org/10.1093/jac/dkt452.
-
27.
Stegger M, Andersen PS, Kearns A, Pichon B, Holmes MA, Edwards G, et al. Rapid detection, differentiation and typing of methicillin-resistant Staphylococcus aureus harbouring either mecA or the new mecA homologue mecA(LGA251). Clin Microbiol Infect. 2012;18(4):395-400. [PubMed ID: 22429460]. https://doi.org/10.1111/j.1469-0691.2011.03715.x.
-
28.
García-Álvarez L, Holden MT, Lindsay H, Webb CR, Brown DF, Curran MD, et al. Meticillin-resistant Staphylococcus aureus with a novel mecA homologue in human and bovine populations in the UK and Denmark: a descriptive study. Lancet Infect Dis. 2011;11(8):595-603. https://doi.org/10.1016/s1473-3099(11)70126-8.
-
29.
Rania AA, Nsreen MK, Rasha HE, Mona MA. Evaluation for the Novel mecC Methicillin Resistance among Methicillin Resistant Staphylococcal Isolates in two Egyptian University Hospitals. Arch Clin Microbiol. 2017;9(71):1-5.