Abstract
Background:
Design and construction of a universal vaccine based on conserved influenza antigens is the best way to protect populations from unforeseen influenza outbreaks. The ectodomain of matrix protein 2 (M2e) and hemagglutinin stalk domain (HA2) are considered as conserved influenza antigens. Genetic linkage of adjuvant HSP70 to these antigens can improve the efficacy of the vaccine.Objectives:
The aim of this study was to produce a chimer protein to confer cross-protection against different subtypes of influenza A virus.Methods:
The chimer form was subjected to in silico modeling and Immunoinformatics prediction analysis. The heat shock protein 70 (HSP70c) gene was cloned into the pET28a vector downstream of 3M2e-HA2 and expressed in Escherichia coli host. The desired chimer protein was purified with the Ni-TED column.Results:
Validation analysis of the tertiary structure model showed that the model is in the range of native conformations. High score epitopes by Immunoinformatics tools were predicted. The expression of purified 3M2e-HA2-HSP70c was confirmed by the western-blotting assay.Conclusions:
The genetic fusion of adjuvant HSP70 to the target antigen may improve the stimulation of immune responses. In silico analysis revealed the appropriate epitopes characterization of conserved antigens. Hence, the constructed chimer protein could be considered as a potential universal vaccine candidate to protect against influenza infection.Keywords
1. Background
Point mutations and genetic re-assortments are two major contributors of genetic diversity in influenza virus, which cause emergent pandemic strains. Traditional influenza vaccines are based on immunity to the surface glycoproteins that are highly variable among different types of influenza viruses; so, an annual reformulation is needed. The best way to protect people against unforeseen pandemic influenza is to develop an improved universal vaccine using most conserved influenza virus antigens (1, 2). The universal influenza vaccine can induce an immune response, which provides not only protection against the homologous virus but also a cross-protection against infections with various subtypes of influenza virus strains, in particular, zoonotic highly pathogenic avian influenza viruses which have been verified to be able to infect humans (3).
Therefore, design and production of epitope-based vaccines using highly conserved regions of influenza viral proteins are considered as a promising way to control and reduce the influenza virus consequences (2). Tompkins et al. found that immunization of mice with M2-based DNA vaccine induced higher levels of M2 specific antibodies production and specific T-cell responses and protected mice from lethal influenza H5N1 virus challenges (4). Despite the highly conserved sequence of M2e among all human influenza A viruses, its immunogenicity and potency are not efficient due to low molecular weight. Therefore, several approaches have been applied to improve the immunogenicity of M2e based universal vaccines. Some of these efforts include using multiple M2e copies to increase the epitope density, combination of other conserved epitopes, and using some various compounds as adjuvants (5-8).
According to a study conducted by Li et al. (2), using an M2e peptide fused to HA2 could improve universal influenza vaccine potency in inducing a cross-reactive immune response against various subtypes of influenza viruses compared to using M2e or HA2 solely. Mice immunized with this novel artificial antigen prompted potent immune response against infection by various subtypes of influenza viruses. The antigenic glycoprotein hemagglutinin is a homotrimer molecule in which each monomer is composed of two subunits, HA1 and HA2. The N-terminal peptide of HA2 contains some highly conserved sequences, especially the first 11 residues named fusion peptide. Fusion peptide is a critical part of hemagglutinin molecule, which took parts in virus-cell fusion (2, 9). Alvarado-Facundo et al. (10), showed that M2 as an ion channel protein was required for the enhancement of HA fusion and these two viral peptides are a co-worker in viral action. Therefore, due to the critical role of these proteins in virus propagation, chimer subunit vaccines containing these proteins are promising candidates for developing a cross-protective universal influenza vaccine.
HSP70 belongs to conserved chaperon families that can activate adaptive and innate immune responses. Hence, induction of antibodies and high T-cell responses would be increased by fusing a molecular adjuvant like HSP70 to the antigen (11, 12). Shokouhi et al. showed that truncated Leishmania major HSP70 peptide fused to 3M2e influenza conserved domain could stimulate humeral and cellular immune responses compared to 3M2e peptide alone (In press).
2. Objectives
In this study, to stimulate broad and high levels of influenza virus immune responses, we designed a novel chimer protein containing truncated HSP70 fused to 3M2e-HA2 chimer antigen, which was constructed in our lab previously in order to introduce a universal influenza vaccine candidate (13).
3. Methods
3.1. Bioinformatics Analysis
3.1.1 Prediction of Physicochemical Properties, Secondary, and Tertiary Structures
Physicochemical properties of chimer protein were determined using ProtParam online tool (http://web.expasy.org/protparam/) (14). Secondary and tertiary structure prediction is a basic step for the understanding of the protein immunogenic behavior. An accurate secondary structure prediction is a bridge to tertiary structure analysis. In this study, the secondary structure was predicted using self-optimized prediction method with alignment (SOPMA) (https://npsa-prabi.ibcp.fr/cgi-bin/npsa_automat.pl?page=/NPSA/npsa_sopma.html) (15). The three dimensional model of the chimer protein was predicted by I-TASSER server (http://zhanglab.ccmb.med.umich.edu/I-TASSER) (16). The predicted model was visualized by the Swiss PDB viewer 4.1 software.
3.1.2. Validation of the Three-Dimensional Model
The C-Score, TM-score, and RMSD of the modeled structure were calculated by I-TASSER server. The modeled structure was validated by using ProSa (https://prosa.services.came.sbg.ac.at/prosa.php) and Ramachandran Plot in RAMPAGE Analysis server (http://mordred.bioc.cam.ac.uk/~rapper/rampage.php) (17, 18). The Z-score was generated using ProSa server.
3.1.3. Prediction of B-Cell Epitope
Continuous B-cell epitopes of the chimer protein were predicted by using ABCpred server (http://crdd.osdd.net/raghava/abcpred/) (19). The prediction of conformational B-cell epitopes from protein three-dimensional structural data was performed using ElliPro server at http://tools.iedb.org/ellipro/ (20).
3.1.4. Prediction of Cytotoxic T Lymphocyte Epitope
CTLPred server (http://crdd.osdd.net/raghava/ctlpred/) was used to predict CTL epitope of 3M2e-HA2-HSP70c (21).
3.2. Polymerase Chain Reaction Amplification of HSP70c
The C-terminal domain of HSP70 gene (accession number L14604) was PCR amplified from the previously constructed pET28a-HSP70 vector template, using upstream 5’-GCAAGCTTGTGTCCGCGGAG-3’ and downstream 5’-CTTCTCGAGGCTCTCCAGCTCC-3’ primers with HindIII and XhoI restriction sites (bold sequences, respectively). The amplification was carried out in a 25-μL total volume containing 1 μL of DNA template, 2.5 μL of 10x reaction buffer, 0.5 μL of dNTPs (10 mM), 0.75 μL of MgCl2 (50 mM), 1 μL of each primer, and one unit of Taq DNA polymerase (Fermentas, Canada). The PCR program used for amplification was performed as follows: an initial denaturation for 5 minutes at 94°C, followed by 35 cycles for 30 seconds at 94°C, annealing for 30 seconds at 57°C, extension for 30 seconds at 72°C, and a final extension for 5 minutes at 72°C. The PCR products were analyzed by using 1.5% agarose gel electrophoresis and quick gel extraction kit (Invitrogen, USA) was used for purification of the PCR products.
3.3. Cloning of HSP70c Fragment Into pGEM-T Easy Vector
The purified PCR production (342 bp) was cloned into the pGEM-T easy vector (Promega, USA) using T4 DNA ligase (Promega, Germany). The construction of pGEM-HSP70c was transformed into the Top10F’ strain of Escherichia coli chemically competent cells. Positive colonies were chosen by colony-PCR using specific primers and enzyme digestion.
3.4. Construction of Recombinant Expression Vector
Recombinant pGEM-HSP70c was digested by HindIII and XhoI (Fermentas, Canada). HSP70c fragment was purified from the gel by a QIAquick gel extraction kit (Qiagen, USA) and ligated by T4 DNA ligase to the same restriction enzymes digested pET28a-3M2e–HA2 recombinant expression vector (Invitrogen, USA), downstream of the 3M2e-HA2, and transformed into Top10F’ competent cells. The transformed cells were cultured on Luria-Bertani (LB) agar plates containing 50 μg/mL of kanamycin. The presence of HSP70c in the recombinant expression vector was confirmed by colony-PCR, restriction enzyme digestion analysis, and DNA sequencing. The confirmed construct was named pET28a-3M2e-HA2-HSP70c. The target chimer protein, 3M2e-HA2-HSP70c, was translated from the ATG start codon to the TGA stop codon, derived from vector sequence, in a unit open reading frame with two His-tag at the carboxyl and amino terminus of the chimer protein.
3.5. Expression of Chimer Protein in the Prokaryotic System
Recombinant expression vector pET28a-3M2e-HA2-HSP70c was purified from the transformed Top10F’ cells by an extraction kit (Bioneer, Korea). The construct was transformed into the BL21 (Novagen, USA) chemically competent cells. A single colony of transformed cells was cultured in 5ml of LB broth containing 50μg/ml kanamycin and incubated at 37°C in shaker incubator until the optical density at 600 nm reached 0.5. After about 2 hours, the optical density reached 0.5 and expression of the target chimer protein was induced with 0.5 mM concentration of isopropyl β-D-1-thiogalactopyranoside (IPTG) in the culture at 37°C.
3.6. SDS-PAGE and Western Blotting
The expression of the chimer protein was analyzed on a 12% SDS-polyacrylamide gel electrophoresis. Then, proteins were stained by using coomassie brilliant blue R-250 and a protein ladder (Fermentas, Germany) was used for detection of protein size. For western blotting, proteins separated by SDS-polyacrylamide gel electrophoresis were transferred onto nitrocellulose paper using a semi-dry transfer system (Bio-Rad). Then, the immunoblotting was performed using horseradish peroxidase (HRP) conjugated anti-His antibody (Qiagen, Germany). Finally, the protein bands were visualized by their exposure to diaminobenzidine (DAB) substrate.
3.7. Purification of 3M2e-HA2-HSP70c Chimer Protein
The chimer protein was purified based on 6xHis-tag at the carboxyl and amino terminus through the application of Ni–NTA affinity column (M.N, Germany). To purify the chimer protein, E. coli BL21 cells were grown and induced in 250 ml of LB broth as mentioned before. The bacterial cell pellet was re-suspended in 10 ml of lysis buffer. The cells were lysed by sonication with 20 pulses at 20 seconds intervals for eight times on the ice. After loading of the lysates on the Ni-TED column and washing steps, desired chimer protein was eluted by using imidazole buffer. The protein was dialyzed thrice against phosphate-buffered saline (PBS), with buffer changes every 2 hours to remove the urea and imidazole. The purity of the chimer protein was electrophoresed on SDS-polyacrylamide gel electrophoresis and confirmed with western blotting assay.
4. Results
4.1. In silico Analysis Outcome
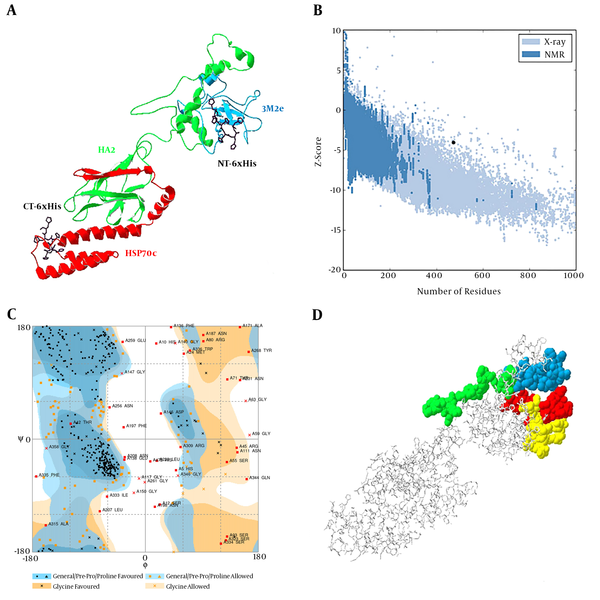
The results of bioinformatics studies such as physicochemical properties by the available software on Expasy website provided information on chimer protein (3M2e-HA2-HSP70c), such as its molecular weight of approximately 52 kDa, the stability index of about 36.13 (below 40 indicates the stability of the molecule), estimated pI (5.44), aliphatic index (6596), and GRAVY (-0.830) (negative GRAVY value indicates that protein is nonpolar). The secondary structure results of chimer protein showed that random coil makes 45.67% while extended strand and alpha helix contribute as 18.6% and 35.73%, respectively.
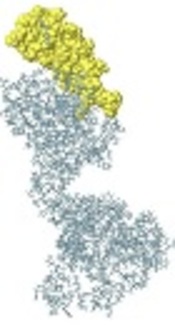
3M2e-HA2-HSP70c chimer protein was modeled using the I-TASSER server and the best-predicted model with the highest confidence score (C-Score - 1.44) was selected for validation analyses (Figure 1A). In addition, the estimated TM-score and estimated RMSD for this model were 0.54 ± 0.15 and 10.6 ± 4.6A, respectively. The structural analysis of the predicted model by ProSa showed that the model is in the range of native conformations of the same size (Figure 1B). Structural evaluation of the three-dimensional model with a total number of 473 amino acids was performed by Ramachandran plot analysis. The evaluation of the plot revealed that 73.2% of the residues are in the favored regions, 18.0% residues in the allowed regions, and 8.7% residues are in the outlier region. The Ramachandran plot provided by RAMPAGE server shows the most residues were fallen in the favorable regions (91.2%) (Figure 1C). Therefore, the predicted three-dimensional structure is suitable.
A, predicted model of chimer protein shown as line ribbon. NT-His and CT-His are shown as stick models with carbon atoms in black color; B, The Z-score plot of the best model; C, Ramachandran plot of tertiary model predicted by RAMPAGE; D, B-cell predicted epitopes using ABCpred: 18 - 37, 39 - 58, 153 - 172, and 172 - 191 are shown in yellow, red, green, and blue colors, respectively, as space-filling model.
4.2. B-Cell Epitope Prediction
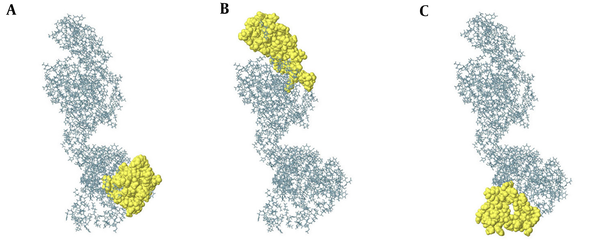
Identification of continuous B-cell epitopes could be useful in epitope-based vaccine design and antibody production. The prediction of the most probable B-cell epitopes in 3M2e-HA2-HSP70c was performed using ABCpred server by setting the cut-off value at > 0.8 and the length of the peptides was fixed as 20-mer. The top scoring peptides were selected as the appropriate epitopes (Table 1) and the location of these predicted epitopes was shown as space-filling model on the surface of the protein (Figure 1D). ElliPro is a powerful web server for the prediction of conformational B-cell epitopes using a three-dimensional structure. The number of residues involved in B-cell epitopes and their scores is given in Table 2. The position of predicted epitopes is demonstrated on the surface protein in Figure 2 (A through C).
List of Predicted Continuous B-cell Epitopes of 3M2e-HA2-HSP70c with the Highest Score Using ABCpred
| Rank | Sequence | Start Position | Score |
|---|---|---|---|
| 1 | HQNEQGSGYAADLKSTQNAI | 153 | 0.93 |
| 2 | IDEITNKVNSVIEKMNTQFT | 172 | 0.92 |
| 3 | GSHMASMTGGQQMGRGSSLL | 18 | 0.90 |
| 4 | EVETPIRNEWGSRSNDSSDG | 39 | 0.89 |
Predicted Conformational B-Cell Epitopes of 3M2e-HA2-HSP70c
Conformational B-cell epitopes of 3M2e-HA2-HSP70c predicted from the modeled structure (A to C). The result was viewed by Jmol viewer. In the ball-and-stick model, yellow balls are the residues of Conformational predicted epitopes and sticks are non-epitope residues structures.
4.3. T-Cell Epitope Prediction
Cytotoxic T-cell epitope regions of 3M2e-HA2-HSP70c chimer protein were predicted using the CTLPred server. The three top predicted CTL epitopes are FNHLEKRIE, EAKLNREEI, and LKSTQNAID with a score of 1, 1, and 0.99, respectively.
4.4. Construction of pET-28a-3M2e-HA2-HSP70c
C-terminal domain of L. major HSP70 was amplified (Figure 3A). The HSP70c gene was successfully ligated into the C-terminal of 3M2e-HA2 to form a 3M2e-HA2-HSP70c fused gene as a unit open reading frame. The recombinant vector was confirmed by colony-PCR (Figure 3B), restriction enzyme digestion (Figure 3C) and sequencing.
A, PCR result of HSP70 fragment amplification on 1.5% agarose gel: lane 1: 100 bp DNA ladder, lane 2: negative control, lane 3 - 4: PCR amplification of target gene; B, colony PCR results, lane1: 100 bp DNA ladder, lane 2: negative control, lane 3 - 6: confirmed colony; C, restriction enzyme analysis of chimer construct, Lane1: 100 bp DNA ladder, lane 2: un cutting construct, lane 3: cutting construct.

4.5. Expression, Purification, and Confirmation of the 3M2e-HA2-HSP70c Chimer Protein
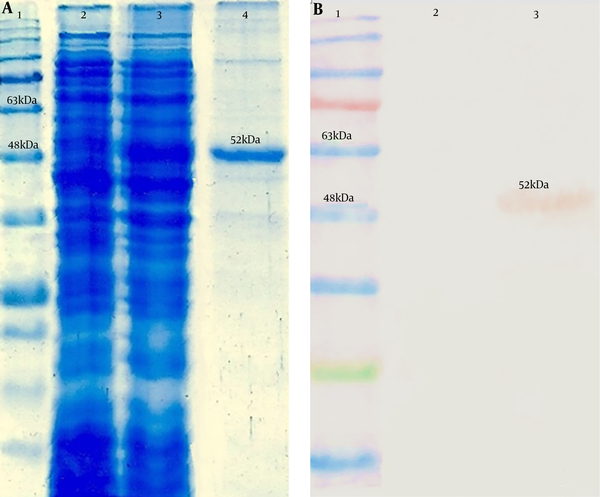
The expressed chimer protein was detected in 12% SDS–PAGE (Figure 4A). Purification of the chimer protein was done and a 52-kDa band corresponding to the purified chimer protein was observed on SDS-PAGE. Confirmation of the chimer protein was performed by western blotting assay through observation of expected band (Figure 4B).
Analysis of the 3M2e-HA2-HSP70c chimer protein expression by SDS-PAGE and western blotting assays. A, Lane 1: Protein ladder (Fermentas), lane 2: Expression before IPTG induction, lane 3: Expression after IPTG induction, lane 4: purified chimer protein; B, Lane 1: Protein ladder (Fermentas), Lane 2: negative control, Lane 3: 6xHis-tag-based purified chimer protein.
5. Discussion
HSPs are the members of the molecular chaperones family. Due to their role in the activation of Antigen-presenting cells, the secretion of proinflammatory cytokines, and dendritic cells maturation, various studies used HSPs as a molecular adjuvant in epitope-based vaccines (22, 23). Thus, HSPs can be used as a carrier molecule for antigens to enhance the immune response (24, 25). In a previous study, fusing C-terminus of mycobacterial HSP70 to four tandem repeats of the conserved influenza matrix protein M2 enhanced the immunogenicity of the antigens and protected mice from a lethal dose of the influenza A virus challenge (11). Herein, we developed a new construct of influenza virus antigens encoding three tandem repeats of the M2e peptide fused to highly conserved stalk domain of hemagglutinin that contained immunogenic and protective epitopes. In this construction, the C-terminal domain of L. major HSP70 (amino acids 491-604) was used as a molecular adjuvant. The entire fragment of HSP70 is a large antigen and thus difficult to use as an epitope-based vaccine in linkage with other peptides. Therefore, it is needed to choose truncated forms of HSP70 that induce a potent immune response. Rafati et al. (26), evaluated the antigenicity effects of L. major HSP70 in an animal model and found that the C-terminal of HSP70 (amino acids 491-604) contained the highest immunogenicity.
For better accessibility of antibody to the highly conserved epitope of an M2 protein, located at N-terminus of M2e, HSP70c gene was fused to the C-terminal of the 3M2e-HA2 construct. Identification of viral conserved regions capable of promoting strong humoral and cellular immune responses is crucial in the development of novel universal vaccines. These selected targeting regions must be conserved among hypervariable influenza viruses. In-silico structure and epitope analysis of universal vaccine candidates is a useful approach for the rational design of epitope-based vaccines. The combination of computational analysis and laboratory techniques can eliminate the time, effort, and cost of vaccine studies. In the present study, 3M2e-HA2-HSP70c chimer protein was analyzed using bioinformatics tools.
In-silico analysis was applied in computational biotechnology for rational verification of chimer protein for influenza vaccine development. Structural analysis of the modeled three-dimensional structure by ProSa server indicated that I-TASSER server could predict a good resolution model with the correct topology. Ramachandran plot analysis of the predicted model using RAMAPAGE indicated that most of the amino acid residues were in the allowed regions. A successful candidate vaccine must be capable of eliciting a protective immune response. Therefore, identification of the most prominent B-cell and T-cell epitopes is a crucial step in epitope-based vaccine design. Prediction of CTL epitopes was performed using CTLPred. ABCpred was used to predict continuous B-cell epitopes. Three-dimensional structure analysis revealed that the continuous B-cell immunodominant epitopes were located on the surface of the chimer protein. Ellipro tool was used to identify the B-cell conformational epitopes with the structure-based algorithm. This algorithm works on three-dimensional structures of the protein.
5.1. Conclusions
Bioinformatics analysis revealed the appropriate epitope characterization of these conserved antigens. In vitro construction, expression, and purification showed that the predicted physicochemical properties such as molecular weight are compatible with in silico data. Experimental validation of 3M2e-HA2-HSP70c construct is among the essential steps toward using predicted conserved epitopes as an effective universal vaccine. Our results suggest that the 3M2e-HA2-HSP70c may induce humoral and cellular immune responses against different types of influenza viruses. Hence, constructed chimer protein could be considered as a potential universal vaccine candidate although its immunogenicity needs to be investigated in animal model among other influenza virus conserved proteins.
Acknowledgements
References
-
1.
Berlanda Scorza F, Tsvetnitsky V, Donnelly JJ. Universal influenza vaccines: Shifting to better vaccines. Vaccine. 2016;34(26):2926-33. [PubMed ID: 27038130]. [PubMed Central ID: PMC4899887]. https://doi.org/10.1016/j.vaccine.2016.03.085.
-
2.
Li J, Arevalo MT, Chen Y, Posadas O, Smith JA, Zeng M. Intranasal immunization with influenza antigens conjugated with cholera toxin subunit B stimulates broad spectrum immunity against influenza viruses. Hum Vaccin Immunother. 2014;10(5):1211-20. [PubMed ID: 24632749]. [PubMed Central ID: PMC4896531]. https://doi.org/10.4161/hv.28407.
-
3.
de Vries RD, Altenburg AF, Rimmelzwaan GF. Universal influenza vaccines: a realistic option? Clin Microbiol Infect. 2016;22 Suppl 5:S120-4. [PubMed ID: 27130671]. https://doi.org/10.1016/j.cmi.2015.12.005.
-
4.
Tompkins SM, Zhao ZS, Lo CY, Misplon JA, Liu T, Ye Z, et al. Matrix protein 2 vaccination and protection against influenza viruses, including subtype H5N1. Emerg Infect Dis. 2007;13(3):426-35. [PubMed ID: 17552096]. [PubMed Central ID: PMC2725899]. https://doi.org/10.3201/eid1303.061125.
-
5.
Jang YH, Seong BL. Options and obstacles for designing a universal influenza vaccine. Viruses. 2014;6(8):3159-80. [PubMed ID: 25196381]. [PubMed Central ID: PMC4147691]. https://doi.org/10.3390/v6083159.
-
6.
Kesik-Brodacka M, Plucienniczak G. A universal flu vaccine. Acta Biochim Pol. 2014;61(3):523-30. [PubMed ID: 25203218].
-
7.
Liu W, Zou P, Ding J, Lu Y, Chen YH. Sequence comparison between the extracellular domain of M2 protein human and avian influenza A virus provides new information for bivalent influenza vaccine design. Microbes Infect. 2005;7(2):171-7. [PubMed ID: 15777646]. https://doi.org/10.1016/j.micinf.2004.10.006.
-
8.
Rappazzo CG, Watkins HC, Guarino CM, Chau A, Lopez JL, DeLisa MP, et al. Recombinant M2e outer membrane vesicle vaccines protect against lethal influenza A challenge in BALB/c mice. Vaccine. 2016;34(10):1252-8. [PubMed ID: 26827663]. https://doi.org/10.1016/j.vaccine.2016.01.028.
-
9.
Sui J, Hwang WC, Perez S, Wei G, Aird D, Chen LM, et al. Structural and functional bases for broad-spectrum neutralization of avian and human influenza A viruses. Nat Struct Mol Biol. 2009;16(3):265-73. [PubMed ID: 19234466]. [PubMed Central ID: PMC2692245]. https://doi.org/10.1038/nsmb.1566.
-
10.
Alvarado-Facundo E, Gao Y, Ribas-Aparicio RM, Jimenez-Alberto A, Weiss CD, Wang W. Influenza virus M2 protein ion channel activity helps to maintain pandemic 2009 H1N1 virus hemagglutinin fusion competence during transport to the cell surface. J Virol. 2015;89(4):1975-85. [PubMed ID: 25473053]. [PubMed Central ID: PMC4338904]. https://doi.org/10.1128/JVI.03253-14.
-
11.
Ebrahimi SM, Dabaghian M, Tebianian M, Jazi MH. In contrast to conventional inactivated influenza vaccines, 4xM2e.HSP70c fusion protein fully protected mice against lethal dose of H1, H3 and H9 influenza A isolates circulating in Iran. Virology. 2012;430(1):63-72. [PubMed ID: 22595444]. https://doi.org/10.1016/j.virol.2012.04.015.
-
12.
Ebrahimi SM, Tebianian M. Heterologous expression, purification and characterization of the influenza A virus M2e gene fused to Mycobacterium tuberculosis HSP70(359-610) in prokaryotic system as a fusion protein. Mol Biol Rep. 2010;37(6):2877-83. [PubMed ID: 19813102]. https://doi.org/10.1007/s11033-009-9846-2.
-
13.
Jalili N, Taheri N, Tavakoli R, Fotoohi F, Akbari A, Farahmand B. [Expression and Purification of a Recombinant Chimeric Protein (3M2e-HA2) Composed of Influenza Virus Hemaglutinin and Matrix Protein Conserved Domain for Universal Subunit Vaccine Development]. J Mazandaran U Med Sci. 2016;26(137):12-22. Persian.
-
14.
Wilkins MR, Gasteiger E, Bairoch A, Sanchez JC, Williams KL, Appel RD, et al. Protein identification and analysis tools in the ExPASy server. Methods Mol Biol. 1999;112:531-52. [PubMed ID: 10027275].
-
15.
Geourjon C, Deleage G. SOPMA: significant improvements in protein secondary structure prediction by consensus prediction from multiple alignments. Comput Appl Biosci. 1995;11(6):681-4. [PubMed ID: 8808585].
-
16.
Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics. 2008;9:40. [PubMed ID: 18215316]. [PubMed Central ID: PMC2245901]. https://doi.org/10.1186/1471-2105-9-40.
-
17.
Lovell SC, Davis IW, Arendall WB, Bakker PI, Word JM, Prisant MG, et al. Structure validation by Calpha geometry: phi,psi and Cbeta deviation. Proteins. 2003;50(3):437-50. [PubMed ID: 12557186]. https://doi.org/10.1002/prot.10286.
-
18.
Wiederstein M, Sippl MJ. ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 2007;35(Web Server issue):W407-10. [PubMed ID: 17517781]. [PubMed Central ID: PMC1933241]. https://doi.org/10.1093/nar/gkm290.
-
19.
Saha S, Raghava GP. Prediction of continuous B-cell epitopes in an antigen using recurrent neural network. Proteins. 2006;65(1):40-8. [PubMed ID: 16894596]. https://doi.org/10.1002/prot.21078.
-
20.
Ponomarenko J, Bui HH, Li W, Fusseder N, Bourne PE, Sette A, et al. ElliPro: a new structure-based tool for the prediction of antibody epitopes. BMC Bioinformatics. 2008;9:514. [PubMed ID: 19055730]. [PubMed Central ID: PMC2607291]. https://doi.org/10.1186/1471-2105-9-514.
-
21.
Bhasin M, Raghava GP. Prediction of CTL epitopes using QM, SVM and ANN techniques. Vaccine. 2004;22(23-24):3195-204. [PubMed ID: 15297074]. https://doi.org/10.1016/j.vaccine.2004.02.005.
-
22.
Ebrahimi SM, Tebianian M. Role of mycobacterial heat shock protein 70 (mHSP70) as genetic vaccine adjuvants. World Appl Sci J. 2011;14(10):1569-75.
-
23.
Li X, Yang X, Li L, Liu H, Liu J. A truncated C-terminal fragment of Mycobacterium tuberculosis HSP70 gene enhanced potency of HBV DNA vaccine. Vaccine. 2006;24(16):3321-31. [PubMed ID: 16472546]. https://doi.org/10.1016/j.vaccine.2006.01.012.
-
24.
Rico AI, Angel SO, Alonso C, Requena JM. Immunostimulatory properties of the Leishmania infantum heat shock proteins HSP70 and HSP83. Mol Immunol. 1999;36(17):1131-9. [PubMed ID: 10698315].
-
25.
Udono H, Yamano T, Kawabata Y, Ueda M, Yui K. Generation of cytotoxic T lymphocytes by MHC class I ligands fused to heat shock cognate protein 70. Int Immunol. 2001;13(10):1233-42. [PubMed ID: 11581168].
-
26.
Rafati S, Gholami E, Hassani N, Ghaemimanesh F, Taslimi Y, Taheri T, et al. Leishmania major heat shock protein 70 (HSP70) is not protective in murine models of cutaneous leishmaniasis and stimulates strong humoral responses in cutaneous and visceral leishmaniasis patients. Vaccine. 2007;25(21):4159-69. [PubMed ID: 17395340]. https://doi.org/10.1016/j.vaccine.2007.03.006.