Abstract
Background:
Ulcerative colitis is a kind of inflammatory bowel disease that is considered as immunological response to commensal bacteria colonizing gut lumen. Adherent-invasive Escherichia coli strains are pathogens responsible for ulcerative colitis disease. These bacteria have special virulence factors, including type 1 fimbriae, which could be involved in inflammatory bowel disease.Objectives:
The present study was conducted to determine the prevalence of adherent-invasive E. coli with fimH gene isolated from Iranian patients with ulcerative colitis.Methods:
Sixty intestinal biopsy samples of 30 patients with ulcerative colitis and 30 individuals without inflammatory bowel disease were examined. Biopsies from rectum, descending, ascending, terminal ileum, and colon were taken during colonoscopy.Results:
All biopsy samples were cultured for isolation of E. coli strains. Using polymerase chain reaction assay, the invasive plasmid antigen H and invasion-association locus genes were detected from both isolated bacteria and tissue specimens to confirm the presence of adherent-invasive E. coli. The frequency of adherent-invasive E. coli with type 1 fimbriae was much higher in patients with ulcerative colitis than control subjects. Among isolated bacteria, type 1 fimbriae of adherent-invasive E. coli were detected in 53.3% and 13.3% of ulcerative colitis patients and control subjects, respectively. In addition, from 60 biopsy samples, type 1 fimbriae were detected in 56.7% of ulcerative colitis patients but in 10% of healthy subjects.Conclusions:
Subjects without inflammatory bowel disease had a high rate of E. coli strains than patients with ulcerative colitis via cultivation detection. We found a high rate of type 1 fimbriae of adherent-invasive E. coli in ulcerative colitis patients by polymerase chain reaction assay. It appears that the presence of adherent-invasive E. coli with type 1 fimbriae in the gastrointestinal tract of patients with ulcerative colitis is more likely than previously supposed.Keywords
Escherichia coli Colitis Ulcerative Inflammatory Bowel Diseases PCR
1. Background
Ulcerative colitis (UC) is known as a chronic immune-mediated disease, causing inflammation and ulcers in the colon (1). The first symptom of active disease is diarrhea mixed with blood. Many parts of the body outside the intestinal tract are affected by ulcerative colitis. In fact, the initiation of UC disease is in intestinal zones with high bacterial counts (2, 3). The pathogenesis of UC is complicated even though its cause is still unknown. Genetic background could be a risk factor for UC disease; however, microbiota and immune system may have roles in the occurrence of the disease (4). A wide range of bacterial species is involved in the inflammation of the colon, including Enterobacteriaceae, especially Escherichia coli (5). Escherichia coli strains living in the gut are not considered to be harmful. However, numerous studies have indicated that the number of adherent-invasive E. coli (AIEC) strains in inflammatory bowel disease (IBD) patients is significantly higher than previously estimated (6).
Adherent invasive E. coli can adhere to epithelial cells and invade cytoplasmic eukaryotic infectious cells owing to type 1 fimbriae (fimH), invasive plasmid antigen H (ipaH), and invasion-association locus (ial). Also, it can replicate into macrophages. Accordingly, it should be considered a separate pathogenic cohort triggering intestinal disease in human. Subsequently, studies proposed that AIEC can be associated with pathogenicity of IBD (7). In adherent-invasive E. coli, the fimH gene, in particular, helps in adhering and colonizing epithelial cells.
The ability to invade intestinal cells is acquired by ipaH and ial genes. The ipaH gene exists in multiple copies located on both chromosomes, and the plasmid is in charge of release in epithelial cells (8). Recently, there are no data about the prevalence of adherent-invasive E. coli in the large bowel of patients with inflammation (9). Adherent invasive E. coli pathotype has been gradually involved in the etiopathogenesis of ulcerative colitis (10). A number of culture-based and molecular-based studies support the theory that adherent-invasive E. coli especially those with fimH gene are a microbiological factor effective in IBD (11-13).
2. Objectives
The main aim of the present study was to determine the prevalence of adherent-invasive E. coli with fimH gene isolated from Iranian patients with ulcerative colitis.
3. Methods
3.1. Patients and Tissue Samples
In the present study, a group of biopsy samples of 60 subjects comprising 30 UC patients (12 males, 18 females, median age 36.9 years, age range 16 - 75 years) and 30 individuals without IBD (14 males, 16 females, median age 48.31 years, age range 19 - 76 years) were taken during colonoscopy. The study was approved by the ethics committee of Tehran University Medical Sciences.
3.2. Microbial Identification
Samples obtained during colonoscopy were transferred immediately into sterile vials containing either thioglycolate broth or saline (Sigma-Aldrich, Hi Media) and stored at -20°C. The biopsy specimens were homogenized and inoculated into Hi Chrome E. coli agar (Sigma-Aldrich, Hi Media) and incubated for 18 - 24 hours at 35 ± 2°C. The bacteria were stored in TSB broth containing 30% glycerol at -70°C until further analysis.
3.3. DNA Extraction from Biopsies for PCR
Biopsies were crushed and DNA was extracted by RTP® Mycobacteria kit (Berlin, Germany).
3.4. DNA Extraction from Culture for PCR
Isolated bacteria were prepared for PCR amplification. The bacterial colonies were harvested and centrifuged. The sediments were suspended in 500 µL of sterile deionized water, and boiled for 10 minutes. After centrifugation of the boiled samples at 19000 g for 5 minutes, the supernatant was used as DNA template in PCR assay (14).
3.5. PCR Assay
All isolates were tested for the occurrence of ipaH, ial, and fimH genes using PCR. The nucleotide sequence of primers (Macrogen, Pishgam) and size of product (base pairs) for amplification of fimH, ipaH, and ial genes are displayed in Table 1 (15, 16). PCR was performed in 25 μL solution comprising 5 μL of master mix (Amplicon, Pishgam), 1 μL of each primer (10 pm/μL), 2 μL of DNA template (50 ng), and 16 μL of ddH2O. Subsequently, the following thermal cycling conditions were used: 5 minutes at 94°C and 36 cycles of amplification consisting of 30 seconds at 95°C, 30 seconds at 56°C, and 1 minutes at 72°C, with 5 minutes at 72°C for final extension. PCR products were investigated by electrophoresis on a 1% agarose gel in 1X TBE buffer [10.8 g Tris and 5.5 g Boric acid, 0.5 M Na2EDTA (pH 8.0)] (17).
Primers and Annealing Temperature for Amplification of the AIEC Genes
| Primers | Nucleotide Sequences (5’ - 3’) | Size of Product, bp |
|---|---|---|
| fimH | TATGGCGGCGTGTTATCTAG | 150 |
| CACAGGCGTCAAATAAAGCG | ||
| ipaH | GTTCCTTGACCGCCTTTCCGATACCGTC | 619 |
| GCCGGTCAGCCACCCTCTGAGAGTAC | ||
| ial | CTGGATGGTATGGTGAGG | 320 |
| GGAGGCCAACAATTATTTCC |
3.6. Statistical Analysis
The data were evaluated by Pearson Chi-Square test. A P value < 0.05 was considered statistically significant.
4. Results
Escherichia coli strains were detected from biopsy samples of 24 patients with UC (80%) and 26 healthy controls (86.7%). All isolated bacteria were confirmed as AIEC by biochemical tests and PCR assay. The ipaH, ial, and fimH genes were amplified by utilizing particular primers and became visible at approximate bands of 619, 320, and 150 bp on polyacrylamide gel, respectively (Figures 1, 2 and 3). Among isolated bacteria, the presence of fimH was confirmed in 53.3% (n = 16) of specimens from UC patients, while 46.7% (n = 14) were negative. Also, this gene was detected in 4 control subjects (13.3%) while the remaining 86.7% (n = 26) lacked this gene (Table 2). In addition, amongst 60 biopsy samples taken during colonoscopy, the fimH gene was detected in 17 (56.7%) patients with UC and 3 (10%) control subjects (Figure 3). Accordingly, 43.3% (n = 13) of UC patients and 90% (n = 27) of control subjects did not yield any amplicon in PCR assay (Table 3). Therefore, fimH gene in E. coli strains isolated from UC patients was more frequent than that of control population in PCR assay. Moreover, PCR assay was more reliable than cultivation. Based on the results, the association of fimH gene presence in adherent-invasive E. coli with UC patients was statistically significant (P < 0.05). Also, all positive amplified fragments were sequenced.
Polymerase Chain Reaction Amplification of the ipaH Gene
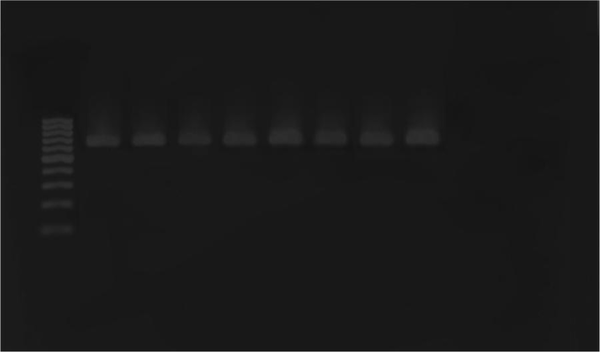
Polymerase Chain Reaction Amplification of the ial Gene
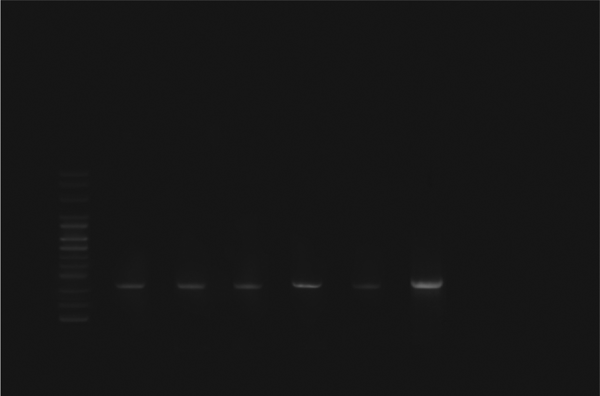
Polymerase Chain Reaction Amplification of the fimH Gene

The Presence of the fimH Gene of AIEC in Isolated from Two Groups (UC Patients and Control Subjects) by PCR Assay
| Population | fimH | ||
|---|---|---|---|
| Positive, No.% | Negative, No.% | P Value | |
| UC, No. = 30 | 16 (53.3%) | 14 (46.7%) | 0.001 |
| Control, No. = 30 | 4 (13.3%) | 26 (86.7%) | 0.001 |
The Presence of the fimH Gene of AIEC in Biopsy Samples from Two Groups (UC Patients and Control Populations) by PCR Assay
| Population | fimH | ||
|---|---|---|---|
| Positive, N.% | Negative, N.% | P Value | |
| UC, No. = 30 | 17 (56.7%) | 13 (43.3%) | 0.000 |
| Control, No. = 30 | 3 (10.0%) | 27 (90.0%) | 0.000 |
5. Discussion
The link between ulcerative colitis disease and the presence of adherent-invasive E. coli carrying fimH gene has been studied in recent years. The pattern of epidemiological characteristics and risk factors related to geographical and regional variations are still unknown (18-20). This study was carried out to determine the prevalence and possible role of AIEC with fimH gene in Iranian patients with UC by PCR assay since limited data are currently available on this subject. To clarify the possible colonization of AIEC in intestinal tract, we screened tissue samples for ipaH and ial genes by PCR assay as these genes have been reported in many cases to be involve in AIEC invasiveness (21). However, there are other studies proposing that these genes are significant virulence determinants in many extra intestinal infections in human, especially infections of urinary tract caused by E. coli (22).
Most studies indicate that modified microbial components and function in IBD can lead to the increased immune response. However, genetic, phenotyping, and microbial diversity within UC patients indicate that this disease is the heterogeneous cohort of different disorders, which probably has predictable natural histories (23). However, recent studies demonstrated that Entrobacteriaceae, E. coli, and Mycobacterium paratuberculosis strains are associated with IBD (24). The current study showed that there was a higher rate of fimH gene in UC patients than control subjects; a difference that was statistically significant. Despite the results of this study, some investigators did not observe any increase in AIEC among UC patients (25, 26).
In this regard, Raso et al. reported that AIEC was not detected in patients with UC and control subjects (27) and likewise, other studies found a higher prevalence for fimbriae I, encoded by the gene fimA, in UC patients compared to healthy individuals (28, 29). However, in our study, 56.7% of the patients with UC were positive for AIEC, in particular for those with fimHgen; a rate that was significantly higher than the rates reported by previous studies. Martin et al. indicated that E. coli is more common in patients with IBD in comparison with control people (30). It can be inferred that fimH gene of AIEC may trigger UC disease. But the absence of fimH gene in AIEC isolated from UC patients may indicate the point that other bacterial genera may also involve in this disease.
In many cases, several studies indicated that invasive E. coli strains are associated with Crohn’s disease (31). Based on previous studies, AIEC strains were identified as true invasive strains involved in the pathogenesis of IBD. The invasive ability of AIEC, acquired by fimH gene, helps its translocation to pass human intestinal barriers and move inside deep tissues (32, 33). In spite of the interesting debate, with the exception of assessing AIEC strains with fimH gene, this gene has been obtained in other strains of E. coli. It can be present in 68% of uropathogenic E. coli (UPEC) strains which are pathogenic in urinary tract (34). The evidence gathered proposes that adhesive and invasive E. coli strains could be involved in ulcerative colitis pathogenesis.
Also, limited data concerning the relation of adherent-invasive E. coli and fimHgen with UC disease in Iran are available. Therefore, we evaluated the presence of fimH gene of AIEC and its relation with UC disease. This study suggests that there is a possible role for this bacterium in pathogenesis of UC disease.
5.1. Conclusion
Several factors have been implicated in the progression of inflammatory bowel disease. In the present study, detection of AIEC with fimH gene from UC patients demonstrated that AIEC with fimH may be a predisposing factor in UC patients. This study provides the first clinical investigation about the relationship between AEIC, especially those with fimH gene, with ulcerative colitis disease in Tehran, Iran.
Acknowledgements
References
-
1.
Wanderas MH, Moum BA, Hoivik ML, Hovde O. Predictive factors for a severe clinical course in ulcerative colitis: Results from population-based studies. World J Gastrointest Pharmacol Ther. 2016;7(2):235-41. [PubMed ID: 27158539]. https://doi.org/10.4292/wjgpt.v7.i2.235.
-
2.
Dignass AU, Baumgart DC, Sturm A. Review article: the aetiopathogenesis of inflammatory bowel disease--immunology and repair mechanisms. Aliment Pharmacol Ther. 2004;20 Suppl 4:9-17. [PubMed ID: 15352888]. https://doi.org/10.1111/j.1365-2036.2004.02047.x.
-
3.
Macfarlane S, Steed H, Macfarlane GT. Intestinal bacteria and inflammatory bowel disease. Crit Rev Clin Lab Sci. 2009;46(1):25-54. [PubMed ID: 19107650]. https://doi.org/10.1080/10408360802485792.
-
4.
Akiho H, Yokoyama A, Abe S, Nakazono Y, Murakami M, Otsuka Y, et al. Promising biological therapies for ulcerative colitis: A review of the literature. World J Gastrointest Pathophysiol. 2015;6(4):219-27. [PubMed ID: 26600980]. https://doi.org/10.4291/wjgp.v6.i4.219.
-
5.
Guarner F, Malagelada JR. Role of bacteria in experimental colitis. Best Pract Res Clin Gastroenterol. 2003;17(5):793-804. [PubMed ID: 14507589].
-
6.
Boudeau J, Glasser AL, Masseret E, Joly B, Darfeuille-Michaud A. Invasive ability of an Escherichia coli strain isolated from the ileal mucosa of a patient with Crohn's disease. Infect Immun. 1999;67(9):4499-509. [PubMed ID: 10456892].
-
7.
Barnich N, Darfeuille-Michaud A. Adherent-invasive Escherichia coli and Crohn's disease. Curr Opin Gastroenterol. 2007;23(1):16-20. [PubMed ID: 17133079]. https://doi.org/10.1097/MOG.0b013e3280105a38.
-
8.
Sousa MA, Mendes EN, Collares GB, Peret-Filho LA, Penna FJ, Magalhaes PP. Shigella in Brazilian children with acute diarrhoea: prevalence, antimicrobial resistance and virulence genes. Mem Inst Oswaldo Cruz. 2013;108(1):30-5. [PubMed ID: 23440111].
-
9.
Willing B, Halfvarson J, Dicksved J, Rosenquist M, Jarnerot G, Engstrand L, et al. Twin studies reveal specific imbalances in the mucosa-associated microbiota of patients with ileal Crohn's disease. Inflamm Bowel Dis. 2009;15(5):653-60. [PubMed ID: 19023901]. https://doi.org/10.1002/ibd.20783.
-
10.
Baumgart M, Dogan B, Rishniw M, Weitzman G, Bosworth B, Yantiss R, et al. Culture independent analysis of ileal mucosa reveals a selective increase in invasive Escherichia coli of novel phylogeny relative to depletion of Clostridiales in Crohn's disease involving the ileum. ISME J. 2007;1(5):403-18. [PubMed ID: 18043660]. https://doi.org/10.1038/ismej.2007.52.
-
11.
Darfeuille-Michaud A, Neut C, Barnich N, Lederman E, Di Martino P, Desreumaux P, et al. Presence of adherent Escherichia coli strains in ileal mucosa of patients with Crohn's disease. Gastroenterology. 1998;115(6):1405-13. [PubMed ID: 9834268].
-
12.
Fujita H, Eishi Y, Ishige I, Saitoh K, Takizawa T, Arima T, et al. Quantitative analysis of bacterial DNA from Mycobacteria spp., Bacteroides vulgatus, and Escherichia coli in tissue samples from patients with inflammatory bowel diseases. J Gastroenterol. 2002;37(7):509-16. [PubMed ID: 12162408]. https://doi.org/10.1007/s005350200079.
-
13.
Gophna U, Sommerfeld K, Gophna S, Doolittle WF, Veldhuyzen van Zanten SJ. Differences between tissue-associated intestinal microfloras of patients with Crohn's disease and ulcerative colitis. J Clin Microbiol. 2006;44(11):4136-41. [PubMed ID: 16988016]. https://doi.org/10.1128/JCM.01004-06.
-
14.
Hojati Z, Zamanzad B, Hashemzadeh M, Molaie R, Gholipour A. The FimH Gene in Uropathogenic Escherichia coli Strains Isolated From Patients With Urinary Tract Infection. Jundishapur J Microbiol. 2015;8(2):17520. [PubMed ID: 25825648]. https://doi.org/10.5812/jjm.17520.
-
15.
Nguyen TV, Le Van P, Le Huy C, Gia KN, Weintraub A. Detection and characterization of diarrheagenic Escherichia coli from young children in Hanoi, Vietnam. J Clin Microbiol. 2005;43(2):755-60. [PubMed ID: 15695676]. https://doi.org/10.1128/JCM.43.2.755-760.2005.
-
16.
da Cruz CB, de Souza MC, Serra PT, Santos I, Balieiro A, Pieri FA, et al. Virulence factors associated with pediatric shigellosis in Brazilian Amazon. Biomed Res Int. 2014;2014:539697. [PubMed ID: 24877110]. https://doi.org/10.1155/2014/539697.
-
17.
Zhang Y, Rowehl L, Krumsiek JM, Orner EP, Shaikh N, Tarr PI, et al. Correction: Identification of Candidate Adherent-Invasive E. coli Signature Transcripts by Genomic/Transcriptomic Analysis. PLoS One. 2015;10(7):134759. [PubMed ID: 26218289]. https://doi.org/10.1371/journal.pone.0134759.
-
18.
Abraham SN, Sun D, Dale JB, Beachey EH. Conservation of the D-mannose-adhesion protein among type 1 fimbriated members of the family Enterobacteriaceae. Nature. 1988;336(6200):682-4. [PubMed ID: 2904657]. https://doi.org/10.1038/336682a0.
-
19.
Klemm P, Christiansen G. Three fim genes required for the regulation of length and mediation of adhesion of Escherichia coli type 1 fimbriae. Mol Gen Genet. 1987;208(3):439-45. [PubMed ID: 2890081].
-
20.
Krogfelt KA, Bergmans H, Klemm P. Direct evidence that the FimH protein is the mannose-specific adhesin of Escherichia coli type 1 fimbriae. Infect Immun. 1990;58(6):1995-8. [PubMed ID: 1971261].
-
21.
Vermeire S, Van Assche G, Rutgeerts P. Laboratory markers in IBD: useful, magic, or unnecessary toys? Gut. 2006;55(3):426-31. [PubMed ID: 16474109]. https://doi.org/10.1136/gut.2005.069476.
-
22.
Mitra A, Palaniyandi S, Herren CD, Zhu X, Mukhopadhyay S. Pleiotropic roles of uvrY on biofilm formation, motility and virulence in uropathogenic Escherichia coli CFT073. PLoS One. 2013;8(2):55492. [PubMed ID: 23383333]. https://doi.org/10.1371/journal.pone.0055492.
-
23.
Barnich N, Carvalho FA, Glasser AL, Darcha C, Jantscheff P, Allez M, et al. CEACAM6 acts as a receptor for adherent-invasive E. coli, supporting ileal mucosa colonization in Crohn disease. J Clin Invest. 2007;117(6):1566-74. [PubMed ID: 17525800]. https://doi.org/10.1172/JCI30504.
-
24.
Flanagan P, Campbell BJ, Rhodes JM. Bacteria in the pathogenesis of inflammatory bowel disease. Biochem Soc Trans. 2011;39(4):1067-72. [PubMed ID: 21787349]. https://doi.org/10.1042/BST0391067.
-
25.
Florin TH, Paterson EW, Fowler EV, Radford-Smith GL. Clinically active Crohn's disease in the presence of a low C-reactive protein. Scand J Gastroenterol. 2006;41(3):306-11. [PubMed ID: 16497618]. https://doi.org/10.1080/00365520500217118.
-
26.
Jiang XL, Cui HF. An analysis of 10218 ulcerative colitis cases in China. World J Gastroenterol. 2002;8(1):158-61. [PubMed ID: 11833094].
-
27.
Raso T, Crivellaro S, Chirillo MG, Pais P, Gaia E, Savoia D. Analysis of Escherichia coli isolated from patients affected by Crohn's disease. Curr Microbiol. 2011;63(2):131-7. [PubMed ID: 21626145]. https://doi.org/10.1007/s00284-011-9947-8.
-
28.
Gombosova L, Lazurova I, Zakuciova M, Curova K, Kmetova M, Petrasova D, et al. Genes of intestinal Escherichia coli and their relation to the inflammatory activity in patients with ulcerative colitis and Crohn's disease. Folia Microbiol (Praha). 2011;56(5):367-72. [PubMed ID: 21877213]. https://doi.org/10.1007/s12223-011-0051-z.
-
29.
Boudeau J, Barnich N, Darfeuille-Michaud A. Type 1 pili-mediated adherence of Escherichia coli strain LF82 isolated from Crohn's disease is involved in bacterial invasion of intestinal epithelial cells. Mol Microbiol. 2001;39(5):1272-84. [PubMed ID: 11251843].
-
30.
Martin HM, Campbell BJ, Hart CA, Mpofu C, Nayar M, Singh R, et al. Enhanced Escherichia coli adherence and invasion in Crohn's disease and colon cancer. Gastroenterology. 2004;127(1):80-93. [PubMed ID: 15236175].
-
31.
Mitsuyama K, Tomiyasu N, Takaki K, Masuda J, Yamasaki H, Kuwaki K, et al. Interleukin-10 in the pathophysiology of inflammatory bowel disease: increased serum concentrations during the recovery phase. Mediators Inflamm. 2006;2006(6):26875. [PubMed ID: 17392581]. https://doi.org/10.1155/MI/2006/26875.
-
32.
Barnich N, Boudeau J, Claret L, Darfeuille-Michaud A. Regulatory and functional co-operation of flagella and type 1 pili in adhesive and invasive abilities of AIEC strain LF82 isolated from a patient with Crohn's disease. Mol Microbiol. 2003;48(3):781-94. [PubMed ID: 12694621].
-
33.
Sartor RB. Microbial influences in inflammatory bowel diseases. Gastroenterology. 2008;134(2):577-94. [PubMed ID: 18242222]. https://doi.org/10.1053/j.gastro.2007.11.059.
-
34.
Tarchouna M, Ferjani A, Ben-Selma W, Boukadida J. Distribution of uropathogenic virulence genes in Escherichia coli isolated from patients with urinary tract infection. Int J Infect Dis. 2013;17(6):e450-3. [PubMed ID: 23510539]. https://doi.org/10.1016/j.ijid.2013.01.025.