Abstract
Background:
In recent decades bacteriophages have been used as treating agents against some pathogens. Also, bacteriophages could be considered as alternatives of antimicrobial agents in plant protection.Objectives:
This study aimed to isolate a pathogen from potato (Solanum tubersom) tubers (Pseudomonas putida), and of its specific bacteriophage from soil and wastewater.Materials and Methods:
Infected samples of soft rot potato tubers were collected from Flavarjan farms (Isfahan province, Iran) to isolate and identify P. putida by biochemical and molecular methods. Soil and wastewater samples were obtained locally to isolate the bacteriophage attributed to P. putida. Soil suspension was centrifuged and then filtrated by 0.2 micrometer Millipore filter. The wastewater was directly filtrated by 0.2 micrometer filter after centrifugation. After incubation of isolated bacteria together with phage contained solution, plaques were detected in nutrient agar. Subsequently, clearance of P. putida liquid culture incubated by added filtrated bacteriophage was observed. The structure and morphological characteristics of P. putida related bacteriophage was remarked by transmission electron microscopy (TEM).Results:
Isolated strain sda2 was identified as Pseudomonas putida with related accession number HQ423667. The result showed that isolated bacteriophage belonged to Cystoviridae family.Conclusions:
We isolated a new strain of P. putida as well as a novel bacteriophage through which potato disease caused by the bacterium could be treated.Keywords
1. Background
Pseudomonas putida as a symbion bacteria is found in plant rhizosphere that is known as an antagonist against plants pathogens bacteria (1). Some bacteria such as Ralstonia solanacearum and Erwinia sp. cause soft rot disease in potato and Solanaceae family (2). Nevertheless, there are some data that demonstrate oxidase positive Pseudomonas such as P. putida may cause soft rot disease in potato, too (3). Bacteriophages are among the most effective inhibitor agents to control bacterial population and change bacterial genome during transduction (4-7). In recent decades bacteriophages have been used as treatment agents against some human and animal pathogens such as E.coli and gastrointestinal infections (8-10). Also, the use of bacteriophage in the plants may protect them via reduction in plant bacterial disorders (11).
2. Objectives
This study aimed to isolate an infecting bacterium from infected potato (Solanum tubersom), and of its bacteriophage from wastewater and farm soils for possible use in treatment of infected potato or biocontrol of the disease.
3. Materials and Methods
Chemical materials were purchased from Merck and Sigma Companies. DNA extraction kit was provided from Cinnagen (Cat No: DN8115C, Iran). Two primers for 16S rRNA gene manipulation were prepared by TAG Copenhagen A/S, Denmark. Millipore filter was defined as 0.22 µm acetate cellulose filters (Orange scientific, Spanish). Lyophilization instrument (HETOSICC, Denmark) was applied to concentrate phage solution. Sonication of samples was performed by a sonicator (Transonic 660/H, Germany). Transmission electron microscopy (TEM) (Phillips CM10) was employed for fine detection of phage particles.
3.1. Isolation of Pseudomonas putida
Infected potato tubers with soft rot symptom were collected from Flavarjan, Isfahan province, Iran. The tissue surface was sterilized by 70% ethanol and rinsed 4-5 times by sterile distilled water; then tubers were cut into pieces with 5mm dimensions and suspended in test tubes containing 5ml sterile double distilled water. The test tubes were incubated at 37oC for 60 minutes; this bacterial suspension (i.e. suspended potato tubers in water) was placed in an agar medium supplemented by 15 gr/l Agar, 5 gr/l Peptone, 5 gr/l NaCl, 2 gr/l Yeast extract, and 1 gr/l beef extract for 24-48 hours.
3.2. Biochemical Tests For Identification of Isolated P. putida
To identify biochemical characteristics of P. putida, various tests were applied including oxidase, catalase, oxidative/fermentative (O/F), motility, utilization of glucose and trehalose, and lecithin tests, as well as growth occurrence of the bacterium in 41°C and 4°C.
3.3. 16S rRNA Gene Sequencing.
Bacterial genomic DNA obtained from pure culture was extracted by DNA extraction kit (Cinnagen Cat No: DN8115C). This extract was examined by electrophoresis in 1 % agarose gel. For 16S rRNA gene amplification, universal primers including RW01 as forward primer (5' AACTGGAGGAAGGTGGGGAT 3') and DG74 as reverse primer (5' AGGAGGTGATCCAACCGCA 3') were used (12). PCR conditions were set as follows: initial denaturation at 95°C for 5 minutes (1 cycle), denaturation at 94°C for 45 seconds, annealing at 54°C for 30 seconds, extension by synthesized pieces of 16S rRNA genes at 72oC for 45 seconds, in 30 cycles, and final extension at 72° C for 5 minutes.
3.4. Isolation of Bacteriophage
To isolate bacteriophage, wastewater and farm soils were collected from Flavarjan, Isfahan province. In the first stage, the wastewater was centrifuged at 12000 rpm for 10 minutes followed by supernatant filtration by 0.22 µm acetate cellulose filters (Orange scientific). In the second stage, in order to isolate bacteriophage from the soil, 30 – 50 ml of 1 % potassium citrate supplemented by 10 gr/l K3C6H5O7.H20, 44.1 gr/l Na2HPO4.7 H20, 24 gr/l KH2PO4, pH 7 (13) was mixed with 10 grams of soil sample and sited on ice. Samples were sonicated (transonic 660/H) in three one-minute intervals with one minute rest on ice. This suspension, then, was placed at 4º C for 24 hours and centrifuged at 12000 rpm in 10 minute. The supernatant was filtrated by 0.22 µm acetate cellulose filter and stored at 4°C in refrigerator.
3.5. Bacteriophage Enrichment
To enrich isolated bacteriophage, 1% of filtrated bacteriophage suspension was included to 50 ml overnight prepared culture of P. putida , followed by rotation of this combination in a shaker (50 rpm) at 28oC for 24 hours. The medium was filtrated by 0.22 µm acetate cellulose filters (Orange scientific) and stored at 4oC on 1% of glycine as well as on several drops of chloroforms (13).
3.6. Bacteriophages Plaque Assay
The pure culture of P. putida was prepared on nutrient agar (or Muller Hinton agar). After 2 hours, 10 µL of filtrated bacteriophage was added to culture plate and incubated at 28oC for 24 hours.
3.7. Bacteriophage Purification and Concentration
Bacteriophage purification and concentration are usually performed by the use of two methods including CsCl density-gradient for sample ultracentrifugation and PEG 8000 for purification (14, 15). In this study, lyophilization method was used for bacteriophage concentration and purification, instead. Thirty ml of bacteriophage solution in test flask was incubated in lyophilization instrument (HETOSICC, DENMARK), that was reduced finally to 5 ml. The concentrated solution was then passed through 0.22 µm cellulose acetate filters and prepared for electron microscopy staining.
3.8. Electron Microscopy
For bacteriophage staining, phage solution was deposited on carbon coated copper grid for two minutes and stained by 2% (w/v) uranyl acetate, pH 4 - 4.5. The solution was drained through filter paper and phage particles were observed through transmission electron microscopy (TEM) (Phillips CM10).
3.9. Chemicals
Other chemicals were purchased from Merck or Sigma representative companies in Iran.
4. Results
4.1. Biochemical Identification of Isolated P. Putida
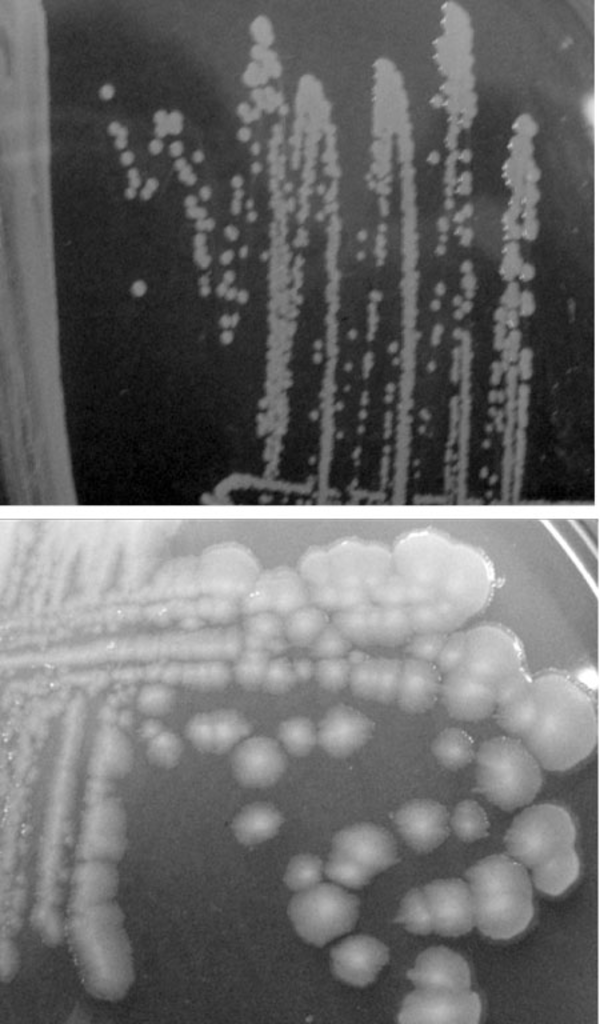
The biochemical and morphological investigations showed that the isolated strain related to P. putida. Positives oxidase and catalase results as well as, positive glucose and negative trehalose utilization, negative growth at 41°C and 4°C, oxidative and non-fermentative (O+/F-) effect, and negative lecithin result were detected. Also, the morphology of isolated bacteria was different from that of other strains of this genus such as P. putida DSM 6125 (Figure 1).
Comparison between Colony Morphology of Standard Pseudomonas putida DSM 6125 (Above) and Isolated P. putida Strain sda2 Extracted From Infected Potato (Solanum tubersom) in This Study (Below).
4.2. 16S rRNA Sequencing
The sequence of 16S rRNA gene PCR product was blasted in NCBI database and mega BLAST program. Data showed that isolated bacterium was attributed to P. putida. Information of bacterial genome was registered in NCBI database as Pseudomonas putida strain sda2 with accession number HQ423667.
4.3. Bacteriophage of P. Putida
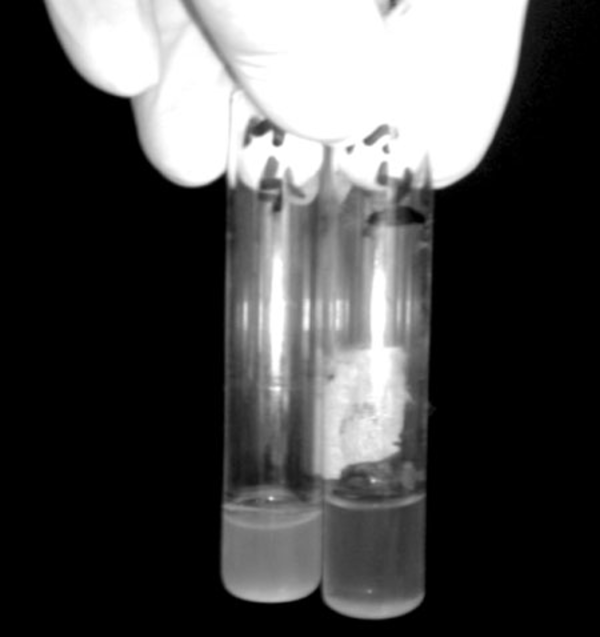
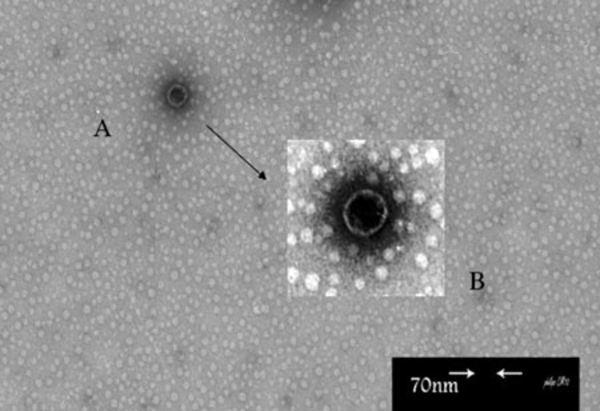
In this study, Broth enrichment method was employed for bacteriophage enrichment. After 10-24 hours, bacterial lysate sedimentation occurred in the tube indicating complete lysis of bacterial cells in the presence of bacteriophage (Figure 2). Small Drop method was used to observe bacteriophage plaques. After bacteriophage inoculation on bacterial culture and incubation for five hours, phage plaques appeared on MHA (Figure 3). Bacteriophage stained by 2% uranyl acetate in pH 4-4.5 was observed in transmission electron microscopy characterized by hexagonal nucleocapsid and about 74 nm across with no tail in its structure indicating that the phage was associated with Cystoviridae family, morphologically (Figure 4). A lipid envelope was detected around the head of bacteriophage.
Complete Lysis of Bacterial Cells (P. putida sda2) In the Presence (Right) and Absence (Left) of Respective Bacteriophage in Liquid Culture After 12 Hours.

Plaque Formation on Pseudomonas putida Strain Sda2 Culture.
Original Electron Micrograph of Bacteriophage Attributed to Pseudomonas putida Strain Sda2.
4.4. Storage of Bacteriophage
Stored bacteriophage samples treated by chloroform were damaged during storage period because of sensitivity to this buffer. However, population of the phage in the samples treated by 1% glycine remained intact.
5. Discussion
In this survey P. putida sda2 was isolated from infected potato tubers. Biochemical tests and 16S rRNA gene homology indicated that isolated strain was attributed to Pseudomonadales family. Morphology of P. putida sda2 colony was different from which of standard strain P. putida DSM 6125. Janse and colleagues have reported that oxidase positive Pseudomonas such as P. putida, P. aureofaciens, P. tolaassii and P. fluorescens have soft rot activity (16). Usually, P.putida does not have any effect on plants and antagonizes Fusarium and Ralstonia effects (17). Shaburva and Krylov have obtained 5 bacteriophages: af, pf16, φ 2f, φ 15, and φ 27 from which φ 15, φ 27, and φ 2f were attributed to Podoviridae family, and phage pf16 to Myoviridae family (18). Lee and Yen studied the effect of bacteriophage gh-1 - isolated by Lee and Boezi in 1966 - on P. putida in oil enhanced recovery. This bacteriophage exhibited a regular hexagonal outline, about 50 µm across, and a short, wedge-shaped tail attached to a corner corresponded to the morphology and size of bacteriophage head and non-contracted tail of Podoviridae family.
In this study, the bacteriophage observed under TEM was characterized by a hexagonal nucleocapsid, about 74 nm across, no visible tail, and being similar to Cystoviridae family in respect of morphology and size. In previous studies, two types of bacteriophages from Podoviridae and Myoviridae families were reported for P. putida but not from Cystoviridae family. Based on international committee on taxonomy of viruses (ICTV) only one bacteriophage (named Phage φ6) was reported to act against Pseudomonas syringae, but there was no report about P. putida.
The cystoviridae family consists of icosahedral enveloped multilayer phages with segmented dsRNA. Our experiments demonstrated that the glycine buffer is a suitable medium to preserve bacteriophages in refrigerator without any effect on the population. In conclusion, we isolated a new strain of P. putida (accession number HQ423667), and a novel bacteriophage for P. putida from Cystoviridae family. In respect to treatment application of bacteriophages against some human and animal pathogens in recent decades, we suggest that phages isolated from natural habitat of plants against infectious agents might be employed to control plant diseases in the same area. Experiments to determine the effectiveness of phage against potato infections should be performed in order to develop a phage control treatment for the disease.
Acknowledgements
References
-
1.
Marugg JD, van Spanje M, Hoekstra WP, Schippers B, Weisbeek PJ. Isolation and analysis of genes involved in siderophore biosynthesis in plant-growth-stimulating Pseudomonas putida WCS358. J Bacteriol. 1985;164(2):563-70. [PubMed ID: 2997118].
-
2.
Dickey RS. Erwinia chrysanthemi: a comparative study of phenotypic properties of strains from several hosts and other Erwinia species. Phytopathology. 1979;69(4):324. https://doi.org/10.1094/Phyto-69-324.
-
3.
Janse J, Derks J, Spit B, Van Der Tuin W. Classification of Fluorescent Soft Rot Pseudomonas Bacteria, Including P. marginalisStrains, Using Whole Cell Fatty Acid Analysis. Syst Appl Microbiol. 1992;15(4):538-53. https://doi.org/10.1016/S0723-2020(11)80114-1.
-
4.
Fuhrman JA. Marine viruses and their biogeochemical and ecological effects. Nature. 1999;399(6736):541-8. [PubMed ID: 10376593]. https://doi.org/10.1038/21119.
-
5.
Fuhrman JA, Schwalbach M. Viral influence on aquatic bacterial communities. Biol Bull. 2003;204(2):192-5. [PubMed ID: 12700152]. https://doi.org/10.2307/1543557.
-
6.
Miao EA, Miller SI. Bacteriophages in the evolution of pathogen-host interactions. Proc Natl Acad Sci U S A. 1999;96(17):9452-4. https://doi.org/10.1073/pnas.96.17.9452.
-
7.
Weinbauer MG, Rassoulzadegan F. Are viruses driving microbial diversification and diversity? Environ Microbiol. 2004;6(1):1-11. https://doi.org/10.1046/j.1462-2920.2003.00539.x.
-
8.
Gorski A, Weber-Dabrowska B. The potential role of endogenous bacteriophages in controlling invading pathogens. Cell Mol Life Sci. 2005;62(5):511-9. [PubMed ID: 15747058]. https://doi.org/10.1007/s00018-004-4403-6.
-
9.
Smith HW, Huggins MB, Shaw KM. The control of experimental Escherichia coli diarrhoea in calves by means of bacteriophages. J Gen Microbiol. 1987;133(5):1111-26. [PubMed ID: 3309177].
-
10.
Verthe K, Possemiers S, Boon N, Vaneechoutte M, Verstraete W. Stability and activity of an Enterobacter aerogenes-specific bacteriophage under simulated gastro-intestinal conditions. Appl Microbiol Biotechnol. 2004;65(4):465-72. [PubMed ID: 14991251]. https://doi.org/10.1007/s00253-004-1585-7.
-
11.
Jones JB, Jackson LE, Balogh B, Obradovic A, Iriarte FB, Momol MT. Bacteriophages for plant disease control. Annu Rev Phytopathol. 2007;45:245-62. [PubMed ID: 17386003]. https://doi.org/10.1146/annurev.phyto.45.062806.094411.
-
12.
Leong D, Greisen K. PCR detection of bacteria found in cerebrospinal fluid. In: Persing DH, editor. Diagnostic Molecular Microbiology: Principles and Applications. Washington: American Society for Microbiology; 1993.
-
13.
Clokie M, Kropinski A. Bacteriophages: methods and protocols, volume 1: isolation, characterization, and interactions. . New York: Humana; 2009.
-
14.
Gnanamanickam SS. Plant-Associated Bacteria. . Dordrecht: Springer; 2006. https://doi.org/10.1007/978-1-4020-4538-7.
-
15.
Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press. 2001.
-
16.
Shaburova OV, Krylov SV, Veiko VP, Pleteneva EA, Burkal'tseva MV, Miroshnokov KA, et al. [Search for destruction factors of bacterial biofilms: comparison of phage properties in a group of Pseudomonas putida bacteriophages and specificity of their halo-formation products]. Genetika. 2009;45(2):185-95. [PubMed ID: 19334612].
-
17.
Chang PL, Yen TF. Interaction of Pseudomonas putida ATCC 12633 and Bacteriophage gh-1 in Berea Sandstone Rock. Appl Environ Microbiol. 1985;50(6):1545-7. [PubMed ID: 16346956].
-
18.
Lee LF, Boezi JA. Characterization of bacteriophage gh-1 for Pseudomonas putida. J Bacteriol. 1966;92(6):1821-7. [PubMed ID: 5958111].