Abstract
Background:
Background: The Lactobacilli belong to lactic acid bacteria, whose primary fermentation end product from sugars is lactic acid and that is why foods are conserved. Lactic acid bacteria have been used for millennia in the production of silage. Therefore, they are an indispensable part of intestinal microflora in human and animals.Objectives:
This research meant to isolate lactic acid bacteria with significant effects from different environments.Materials and Methods:
In this study, heterofermentative LAB were isolated from cheese, yoghurt and corn silage in Broujerd ,Iran. The standard biochemical methods were applied. Acid tolerance was studied by exposure to acidic PBS and growth in bile salt was measured by the spectrophotometric method. The isolated bacteria were studied for antagonistic effects on environment isolated E.coli, bacteriocin and biosurfactant production. Bacterial DNA was extracted, and amplified by PCR method.Results:
The 3 isolates from cheese, yoghurt and silage were effective against isolated E.coli and could produce biosurfactants. Phylogenic relationships of the 3 potential candidates were determined comparing the 16Sr DNA gene sequences, they were found to be as 3 isolates of Lactobacillus buchneri, L.brevis and L.kefiri that were effective on the isolated E.coli from environment.Conclusions:
It was found that the isolated bacteria produced biosurfactants that had a great potential for different industries.Keywords
Lactobacillus Bacteriocins Biosurfactant DNA RNA Ribosomal 16s
1. Background
Lactobacilli are ubiquitous and widespread commensal bacteria in the human and animal microflora. They are widely used by humans: as adjuvants against gastrointestinal disorders, as dietary supplements, and as biological food processors based on their fermentative properties (1). Nucleotide base sequences of Lactobacillus spp. 16S ribosomal DNA (rDNA) provide accurate basis for phylogenetic identification and analysis (2). Lactobacilli are gram-positive, non-spore-forming rods (ranging from coccobacilli to long, slender bacilli). Bacteriocins are proteinaceous antibacterial compounds that exhibit bactericidal activity against species closely related to the producer strain. Several important types of bacteriocins such as nisin, diplococcin, acidophilin, and bulgaricin (3) that are taken from food-associated lactic acid bacteria, have been previously identified and characterized.
Many microorganisms produce extracellular or membrane-associated surface active compounds that play essential roles in the survival of producing microorganisms. Biosurfactants are categorized mainly by their chemical composition and microbial origin. Generally, their structures include a hydrophilic moiety consisting of amino acids or peptides, mono-, di- or polysaccharides and hydrophobic moiety comprising unsaturated or saturated fatty acids. Accordingly, the major classes of biosurfactants include glycolipids, lipopeptides, lipoprotein, phospholipids, fatty acids, polymeric biosurfactants and particulate biosurfactants (4).
2. Objectives
The aim of this research was to determine the strains of Lactobacillus spp. isolated from yoghurt, cheese and silage and also to demonstrate the antimicrobial activity of these isolates.
3. Materials and Methods
3.1. Isolation and Identification
A 10gr sample of each item (cheese, yoghurt and silage) was taken, aseptically. and transferred to the separate sterile plastic bags . Each sample was homogenized in 90 ml of sterile saline solution (0.85%, pH 7). Five 5-fold dilutions of the homogenates were prepared and inoculated on plates of MRS agar and then were incubated anaerobically for 48 hours at 32°C (5, 6). Colonies with typical characteristics were randomly selected from plates and tested for Gram stain, cell morphology, growth at 10°C and 50°C in MRS broth and arginine hydrolysis, catalase and oxidase reaction before further sugar fermentation, growth at 8 and 15 ºC in tubes containing MRS broth, growth in 7.5% NaCl, and fermentation of carbohydrates were determined as described by Sneath et al. (6).
The tested carbohydrates were D (+) cellobiose (Sigma, Detroit, MI, USA), D (+) galactose (Sigma), inulin (Sigma), lactose (Sigma), fructose (Sigma), maltose l-hydrate (Sigma), D mannitol (Sigma), D (+) melezitose (Sigma), melibiose (Sigma), D (-) raffinose (Difco), rhamnose (Sigma), ribose (Sigma), sorbitol (Sigma), D (+) trehalose (Sigma), and D (+) xylose (Merck, Darmstadt, Germany);glucose (Sigma), and sterile water were used as positive and negative controls. During the test, the - isolates were kept in MRS agar stabs at refrigeration temperature. For arginine hydrolysis test, base MRS broth without glucose and meat extract containing 0.3% arginine and 0.2% sodium citrate instead of ammonium citrate was used (1, 7).
3.2. Genetic Analysis of Superior Isolates
3.2.1. Extraction of DNA
DNA was extracted from the bacteria by SET buffer method Signoretto (3). The bacterial pellets were suspended in 1 ml of SET buffer (20% sucrose, 50 mM EDTA, 50 mM Tris-HCl [pH 7.6] containing 1 U of RNase inhibitor, and then 35 ml of lysozyme (5 mg/ml) was added. The suspensions were left on ice for 15 minutes. After this incubation, 9 ml of 25% sodium dodecyl sulfate was added immediately, and samples were incubated for 60 min at room temperature. Subsequently, 50 ml of proteinase K (20 mg/ml) was added, and the suspensions were incubated at room temperature for 60 min, then 0.5 ml ammonium acetate (7.5 m) was added and the suspensions centrifuged at 13000 rpm for 12 min. Two volumes of isopropanol were added to the suspensions and then centrifuged at 13000 rpm for 12 min. The DNA concentration was determined spectrophotometrically. Then, to remove contaminating RNA, 50 µl of the sample was treated with two microliter of DNase - free RNase (3, 8, 9). Partial sequencing of 16S rRNA of two superior isolates was carried out by universal primers:
Forward: 5-CCTACGGGAGGCAGCAG -3, Reverse: 5 GACGTCRTCCNCDCCTTCCT–3.
3.2.2. PCR
The amplification reactions of the DNA sample were carried out in 0.2 mL PCR single tube with hinged flat cap in an eppendorf. Each PCR consisted 5.0 microliter of 10X buffer (10 mM Tris-HCl, 50 mM KCl) +MgSO4 (50 mM) , 1.0 µl dNTPs (5 mM each), 1.0 µl ( 20 pmoles/ microliter ) of each universal primer, 0.5 microliter of pfu DNA polymerase (5 U/µl), 1.0 µl of the DNA template/sample, and sterile water to a volume of 50 µl. The PCR amplification has initial DNA denaturation at 95°C for three min, followed by one cycle of denaturation at 95°C for one min , 30 cycles annealing at 55°C for one min and extension at 72°C for two min, which was followed by a final extension at 72°C for five min. 5 µl PCR product was analyzed by electrophoresis (Bio-Rad) in 1% agarose (Sigma) gel, at 100 volts for 40 min, followed by staining with 1% solution of ethidium bromide (50 µl/L). Gels were visualized by UV trans illumination (3, 8, 9).
Partial sequencing of 16S rRNA of two superior isolates was carried out in Pasteur Institute Laboratories, France. Morphological and biochemical identification tests were also performed following the directions in Bergey’s Systematic Bacteriology Manual (3, 8, 9).
3.3. Evaluation of Antagonistic Activity
Antimicrobial effects of presumptive strains of Lactobacillus spp. against isolated E.coli were determined by the agar diffusion method. The tested bacteria were incubated in nutrient broth at appropriate temperature for 24 hours. Approximately 105- 107 cfu/ml of the bacteria to be tested for sensitivity (indicator bacteria) were inoculated (1%) into 20 ml of nutrient agar and poured into the Petri dishes. To detect antibacterial activity of Lactobacillus spp., MRS containing only 0.2% glucose was used. 10 ml of broth was inoculated with each strain of Lactobacillus spp. and were incubated at 35 ˚C for 48 hours. After incubation, a cell-free solution was obtained by centrifuging (6000 × g for 15 min) the culture, followed by filtration of the supernatant through a 0.2 µm pore size cellulose acetate filter. Some supernatants were neutralized by 1 N NaOH to pH 6.5, and the inhibitory effect of the hydrogen peroxide was eliminated by adding catalase (5 mg/ml). Unneutralized (general inhibitory effect) and neutralized (bacteriocin and bacteriocin-like metabolites) supernatants of the strains of Lactobacillus spp. were checked for antibacterial activity against pathogenic bacteria in inoculated nutrient agar. Then 100 ml of cell free supernatants was filled in 8-mm diameter sealed wells cut in the nutrient agar. Once solidified, the dishes were stored for two hours in a refrigerator. The inoculated plates were incubated for 24 hours at 37 ˚C, and the diameter of the inhibition zone was measured by calipers in millimeters (4, 10-12).
3.3.1. Bacteriocin Detection
Isolated bacteria were propagated in 1000 ml MRS broth (pH 7.0, glucose, 0.25% w/v, peptone, 0.5% w/v) for 72 hours at 37 °C anaerobically (Oxide Gas Generating Kit) in triplicate. To extract bacteriocin, a cell-free solution was obtained by centrifuging (10,000 rpm for 20 min. at 4ºC with Beckman L5050B) the culture and adjusting pH at 7.0 by means of 1M NaOH to exclude the antimicrobial effect of organic acid, followed by filtration of the supernatant through a 0.2 mm pore-size cellulose acetate filter. The supernatant was dialyzed for 24 hours at 4 °C. Inhibitory activity from hydrogen peroxide was eliminated by adding 5 mg/ml catalase (C-100 bovine liver, Sigma) (11). The suspension was boiled for five minutes and the wells were cut in nutrient agar plate by a sterile Pasteur pipette and sealed at the bottom by adding one to two drops of agar. The agar surface was overlaid by isolated E.coli and an additional 30 microliter of the supernatant in the sealed wells and then, the plates were incubated at 37 °C aerobically for 24 hours in triplicate (13, 14).
3.3.2. Preliminary Screening of Biosurfactant Producing Bacteria
A modified oil collapse method was carried out using 96 well microtiter- plates containing 100 µl mineral oil which was equilibrated for an hour at room temperature. 10 µl of supernatant of culture broth was added to the surface of a well and the picture captured after one min using 10 × objective microscope lens. Biosurfactant production was considered positive when the drop diameter was at least 0.5 mm larger than those produced by distilled water and also by culture medium as negative controls (2, 15, 16).
Isolates were screened on blood agar plates containing 5% (v/v) sheep blood and incubated at 37 °C for 48 hours. Hemolytic activity was detected as the presence of a clear zone around bacterial colonies (2, 15, 16).
3.4. Complementary screening
The emulsifying capacity (E24) was evaluated by an emulsification index (E24). The E24 of culture samples was determined by adding 2 ml of kerosene and 2 ml of the cell-free broth in test tube, vortexed at high speed for two minutes and was allowed to stand for 24 hours. The E24 index was given as percentage of the height of emulsified layer (cm) divided by the total height of the liquid column (cm). The percentage of emulsification index was calculated throughthe following equation (2, 15, 16).
Surface tension reduction was measured by Krüss Hamburg Nr 2215 Tensiometer and also by submerging the platinum ring in the cell free culture broth and recording the force required to pull it through the air–liquid interface. The results were compared to that of distilled water and medium composition (as negative control) and Tween 20 (as a positive control). The ability of the isolates to reduce surface tension below 40 mN/m was used as a criterion to select biosurfactant-producing agents (2, 15, 16).
4. Results
4.1. Morphological and Biochemical properties
The isolated bacteria were gram-positive, catalase-negative and heterofermentative bacilli that have yellowish, mocoid, rounded colonies. The carbohydrates fermentation pattern of Lactobacillus spp. has been previously shown.
The isolated bacteria were grown at 10°C and 50°C. These bacteria hydrolyzed arginine and produced NH3. The Production of NH3 was detected by Nesler reagent (Table 1).
Biochemical Tests and Enzyme of Isolates
| Isolates From | |||
|---|---|---|---|
| Cheese | Silage | Yoghurt | |
| Cellobiose | - | - | - |
| Galactose | - | + | + |
| Lactose | + | - | + |
| Fructose | - | - | - |
| Maltose | - | - | + |
| Mannitol | - | - | + |
| Melezitose | - | - | - |
| Melibiose | + | + | - |
| Raffinose | - | - | - |
| Rhamnose | - | + | - |
| Ribose | - | - | + |
| Sorbitol | + | + | - |
| Terehalose | - | + | - |
| Xylose | + | + | - |
| Glucose | + | + | + |
| Arginine dehydrogenase | + | + | + |
| Catalase | + | + | + |
| Oxidase | - | - | - |
| Growth in 7.5% NaCl | + | - | - |
| Growth at 8 ºC | + | + | + |
| Growth at 15 ºC | + | - | - |
4.2. Genetic Analysis of Superior Isolates
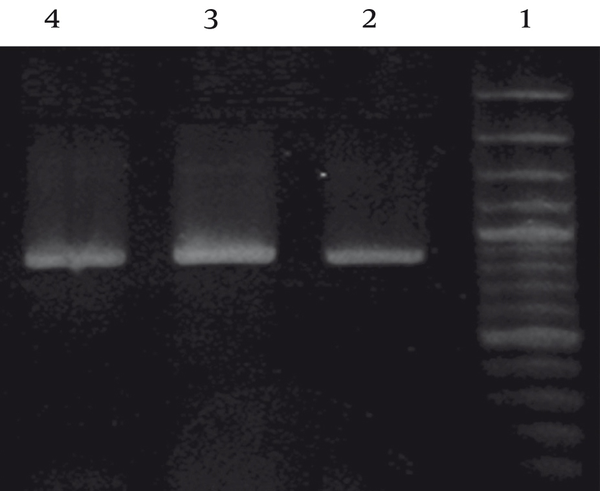
Sequencing 16S rDNA produced by polymerase chain reaction of bacterial DNA using universal primers revealed that superior gram positive biosurfactant-producing isolates were closely related to three Lactobacillus spp., including Lactobacillus buchneri (isolated from yoghurt), L.brevis (isolated from cheese) and L.kefiri (isolated from silage). PCR was performed and products were visualized by agarose gel electrophoresis (Figure 1).
Figure 1. Agarose Gelelectrophoresis of PCR Products
4.3. Bacteriocin and Biosurfactant Production
The isolated bacteria were effective against isolated E.coli from cheese, yoghurt and silage. However, they were not able to produce bacteriocin. The isolates responded positively to oil collapse method and emulsification capacity. Distilled water did not collapse on the oily surface of the well and appeared as a bead. However, the Lactobacillus supernatants collapsed and appeared like firm drops (Figure 2). The putative biosurfactants producing isolates were screened in complementary stage by two methods. Results from these experiments indicate that the surface tension varies from 23.3 mN/m to 57.6 mN/m and the emulsion activity ranges from 0 to 100% for different hydrocarbon sources (Table 2).
Figure 2. Distilled Water Drop on the Oily Surface in the Micro Well Plate (W). (It is not Collapsed). Lactobacilli Supernatants on the Oily Surface. They Are Collapsed.
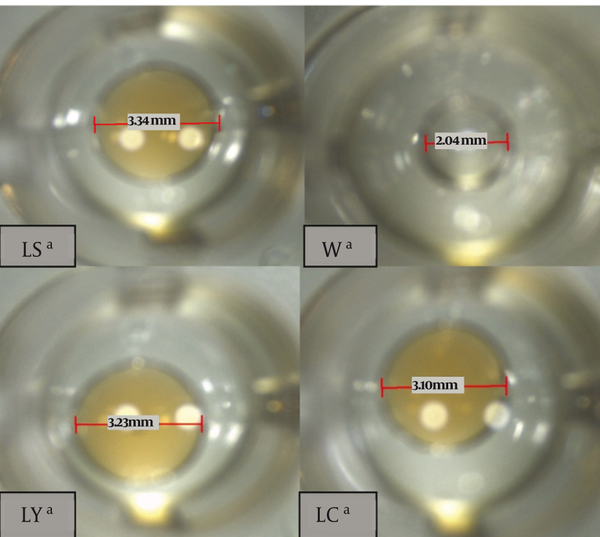
Detection of Biosurfactant Producing Isolates by Preliminary and Complementary Screening Methods.
| Preliminary Methods | Complementary Methods | ||||
|---|---|---|---|---|---|
| Hemolytic Activity | Oil Collapse | Emulsification Capacity (E24) | In Presence of Crude Oil | Surface Tension, mN/m | |
| LC a | + | ++ | 2.27 ± 1.23 | 64 ± 0.56 | 51.6 ± 1.69 |
| LY a | + | ++ | 2.90 ± 0.46 | 72 ± 0.23 | 43.1 ± 0.80 |
| LS a | + | +++ | 2.90 ± 0.46 | 68 ± 1.23 | 57.6 ± 2.29 |
| Water | - | - | 0 ± 0 | 100 ± 0 | - |
5. Discussion
This study tried to screen several types of microorganisms which are capable or incapable of producing biological surfactants or bacteriocin. The drop collapse test was rapid and suitable for this purpose. Results indicated that acid tolerance of isolated Lactobacillus spp. varied at different pHs. Such difference was however lower between isolated Lactobacilli and L. plantarum .The isolated Lactobacilli were bile salt intolerant. In contrast, the isolated bacteria showed significant tolerance to bile salts , however ,when the tolerance level was compared with the standard strain, the difference was insignificant .Pure isolates were cultured in production medium and following centrifugation, supernatants were used to detect bacteriocin and preliminary screening since excretion type bacteria release biosurfactants.
The primary screening of biosurfactants producing bacteria was carried out using hemolytic activity and oil collapse techniques. They did not produce bacteriocin. The results of the current experiments indicated that two isolates were positive for hemolytic activity and none of the isolates were considered negative based on oil collapse and emulsion activities. As already mentioned by Youssef et al. (16) and Plaza et al. (15) in the hemolytic method, there are many bio-products that can cause red blood cell lysis and they do not necessarily have to be surface active molecules.
The drop collapse method is based on the principle that a drop of liquid containing a biosurfactant collapses and spreads over the oily surface. There is a direct relationship between the diameter of the sample and concentration of the biosurfactant and in contrast, the drop lacking biosurfactant remains beaded due to the hydrophobicity of the oil surface that causes aggregation of droplets. Nucleotide base sequences of Lactobacillus spp. 16S ribosomal DNA (rDNA) provide an accurate basis for phylogenetic analysis and identification. The sequences obtained from an isolates can be compared to those of Lactobacillus spp. held in data banks. (8, 9, 11). The 16S region of isolated Lactobacillus spp. have been sequenced and compared with the sequences of type cultures and other valid strains recorded in GenBank (National Center for Biotechnology Information, Bethesda, Md.).
In the present study, isolates (L.buchneri, L.salivarius and L.routeri) with biosurfactant-producing ability were identified. These identified Lactobacillus spp. were in accordance with those earlier identified in different environments by Beasley et al (1). Based on the results of the current study, the employed method is a relatively simple and rapid method by which lactobacilli can be identified without resorting to the use of species-specific PCR primers.
Acknowledgements
References
-
1.
Beasley S. Isolation, identification and exploitation of lactic acid bacteria from human and animal microbiota. Helsinky: University of Helsinki; 2004.
-
2.
Tabatabaee A, Assadi MM, Noohi A, Sajadian V. Isolation of biosurfactant producing bacteria from oil reservoirs. IJEHSE. 2005;2(1).
-
3.
Signoretto C, Lleo MM, Tafi MC, Canepari P. Cell wall chemical composition of Enterococcus faecalis in the viable but nonculturable state. Appl Environ Microbiol. 2000;66(5):1953-9. [PubMed ID: 10788366]. https://doi.org/10.1128/AEM.66.5.1953-1959.2000.
-
4.
Rattanachaikunsopon P, Phumkhachorn P. Lactic acid bacteria: Their antimicrobial compounds and their uses in food production. Ann Biol Res. 2010;1:218-28.
-
5.
Mir-hoseini M. [Study of effect of nisin and producer bacteria of nisin on Listeria monocytogenes and Bacillus cereus]. Isfahan: University of Isfahan; 2004.
-
6.
Sneath PHA, Mair NS, Sharpe ME, Holt JG. Bergey s manual of systematic bacteriology. Baltimore: Williams & Wilkins; 2009.
-
7.
Tannock GW, Tilsala-Timisjarvi A, Rodtong S, Ng J, Munro K, Alatossava T. Identification of Lactobacillus isolates from the gastrointestinal tract, silage, and yoghurt by 16S-23S rRNA gene intergenic spacer region sequence comparisons. Appl Environ Microbiol. 1999;65(9):4264-7. [PubMed ID: 10473450].
-
8.
Boot HJ, Kolen CP, van Noort JM, Pouwels PH. S-layer protein of Lactobacillus acidophilus ATCC 4356: purification, expression in Escherichia coli, and nucleotide sequence of the corresponding gene. J Bacteriol. 1993;175(19):6089-96. [PubMed ID: 8407780].
-
9.
Lleo MM, Tafi MC, Canepari P. Nonculturable Enterococcus faecalis cells are metabolically active and capable of resuming active growth. Syst Appl Microbiol. 1998;21(3):333-9. https://doi.org/10.1016/S0723-2020(98)80041-6.
-
10.
Frizzo L, Zbrun M, Soto L, Bertozzi E, Signorini M, Sequeira G, et al. Protective Effect of an Inoculum of Lactic Acid Bacteria from Bovine Origin Against Salmonella Serotype Dublin in the Intestinal Tract of Mice. J Anim Vet Adv. 2010;9(16):2113-22. https://doi.org/10.3923/javaa.2010.2113.2122.
-
11.
Schurman JJ. Antibacterial activity of hydrogen peroxide against Escherichia coli O157: H7 and Salmonella spp. in fruit juices, both alone and in combination with organic acids. Virginia Polytechnic Institute and State University; 2001.
-
12.
Van Thu T, Foo HL, Loh TC, Bejo MH. Inhibitory activity and organic acid concentrations of metabolite combinations produced by various strains of Lactobacillus plantarum. Afr J Biotechnol. 2011;10(8):1359-63.
-
13.
Ogunbanwo S, Sanni A, Onilude A. Characterization of bacteriocin produced by Lactobacillus plantarum F1 and Lactobacillus brevis OG1. Afr J Biotechno. 2004;2(8):219-27.
-
14.
Todorov SD. Bacteriocin production by Lactobacillus plantarum AMA-K isolated from Amasi, a Zimbabwean fermented milk product and study of the adsorption of bacteriocin AMA-K to Listeria sp. Braz J Microbiol. 2008;39(1):178-87. https://doi.org/10.1590/S1517-83822008000100035.
-
15.
Płaza GA, Zjawiony I, Banat IM. Use of different methods for detection of thermophilic biosurfactant-producing bacteria from hydrocarbon-contaminated and bioremediated soils. J Petrol Sci Eng. 2006;50(1):71-7. https://doi.org/10.1016/j.petrol.2005.10.005.
-
16.
Youssef NH,, Duncan KE, Nagle DP, Savage KN, Knapp RM, McInerney MJ. Comparison of methods to detect biosurfactant production by diverse microorganisms. J Microbiol Methods. 2004;56(3):339-47. [PubMed ID: 14967225]. https://doi.org/10.1016/j.mimet.2003.11.001.