Abstract
Background:
Several epidemic and endemic cases have been reported involving Vibrio cholerae (V. cholerae) as a causative organism in serious diarrheal diseases with high mortality. Hence, quick identification of this microorganism is critical for health care communities.Objectives:
This investigation suggests an accurate, sensitive and rapid test to detect toxigenic and pathogenic species of V. cholerae simultaneously.Materials and methods:
The standard bacteriological tests were utilized to isolate V. cholerae from the clinical samples. A multiplex PCR assay was carried out with three separate primers designed for ctxA, toxR and ompU genes, which play basic roles in toxigenicity and pathogenicity. To investigate the specificity of the designed primers, the amplification statuses of Salmonella typhi (S. typhi), Shigella dysenteriae (S. dysenteriae) and Escherichia coli O157:H7, as common intestinal pathogens, were evaluated. In addition, different concentrations of V. cholerae genome were used for determining the test sensitivity.Results:
From the total of collected stool samples, 72 V. cholerae isolates were found and separated from the patients. All of them carried toxR and ompU genes, but ctxA gene was detected in only 61 isolates. The specificity and sensitivity of this test was 100%.Conclusions:
Obtained data demonstrated that the designed assay is an accurate, easy, rapid, and cost-effective tool for detecting V. cholerae that can serve as an alternate to current bacteriological tests.Keywords
Vibrio cholera Toxin Virulence Gene Polymerase Chain Reaction
1. Background
Diarrheal diseases due to Vibrio cholerae are globally described as one of the most serious problems in a healthy community, even in developed countries. According to WHO reports, more than 100,000 individuals die annually due to cholera diarrhea (1). V. cholerae is a Gram-negative comma-like bacterium. Although 72 species have been categorized in the Vibrio genus, three of them, named V. cholerae, V. parahaemoliticus and V. vulnificus, are the most common ones (2). Reports from the epidemic forms of cholera diarrhea, particularly in developing countries, showes that toxigenic V. cholerae with O1 or O139 sero-group is the main causative microorganism (3).
Cholera toxin, encoded by ctx gene, is a potent factor which induces intense dehydration and pathogenesis (4, 5). Other virulence factors involved in the pathogenecity of V. cholerae include tcp, ompU, and toxR genes (6). OmpU is the most significant outer membrane protein; it can adhere to intestinal epithelium, allowing the bacteria to colonize (7). ToxR is an transcription activating factor that positively regulates expression of several virulent genes, including ctx and toxin co-regulated pilus (TCP) genes and accessory colonization factor (ACF) genes (5, 8, 9). Early identification of this microorganism is essential for each health care community to alleviate the mortality rate. Diagnosis of V. cholerae using medical laboratories routine bacteriological methods is time-consuming. Furthermore, trained and expert technicians are required to perform the detection processes correctly. Since these requirements are not available in some medical laboratories, eliminating these problems with a quick and simple method is desirable (10, 11).
Molecular detection techniques based on PCR are facile and rapid tools with high sensitivity. Real-time PCR (12), loop-mediated isothermal amplification (13), PCR-enzymed linked immunosorbant assay (14) and quadruplex real time PCR (15) are some examples of PCR based methods which have been designed by the researchers. Although the mentioned methods are more sensitive and specific than the conventional PCR, they are more expensive for the patients and occasionally require high level equipment. In resource-limited regions of the world, the majority of laboratories have little availability to perform the described methods due to lack of expert personnel and implement (16). However, the conventional PCR is only capable of detecting one gene in each reaction in addition to being time-consuming. To increase the speed, detection methods based on Multiplex PCR can be useful. Multiplex PCR provides the possibility of evaluating several genes through one PCR reaction in a short time by designing different target-specific primers. This method is simple, fast and cost-saving and has the ability to test a large number of samples at the same time. Moreover, technicians can interpret the results easily (17).
2. Objectives
In this investigation, we designed an improved assay for quick diagnosis of toxigenic and pathogenic V. cholerae with high sensitivity and specificity.
3. Materials and Methods
3.1. Sample Collection
Stool samples were collected during epidemics between 2005 and 2010 from different patients in diverse provinces of Iran.
3.2. Vibrio cholerae Identification
To recognize clinical samples containing V. cholerae strains, the samples were enriched using alkaline peptone water (Merck, Germany) and then cultured on thiosulfate-citrate-bile-sucrose agar (Merck, Germany). All the samples that produced yellow colonies were analyzed further using specific biochemical tests including oxidase testing, sucrose, mannose and arabinose fermentation tests, motility status, indol test, methyl red Voges-Proskauer test, and resistance to polymyxin B. Then the serogroups of clinical V. cholerae isolates were determined by their growth patterns on triple sugar iron agar slants (Merck, Germany) as well as the results of slide agglutination test in the presence of polyvalent O1 and O139 Inaba and Ogawa antisera (Mast Diagnostics Ltd., Bootle, Mersey side, UK) (18). Confirmed V. cholerae strains were stored in luria-Bertani (LB) broth (Merck, Germany) with 30% glycerol and pearl glass at -80°C for further molecular assessments.
3.3. DNA Extraction
In order to perform the molecular experiment, screened isolates were cultured in LB broth for 24 hours at 37°C; then the DNA genome of each one was purified using a DNA extraction kit according to manufacturer’s instructions (Bionner Co, Korea). Quantity and quality of extracted DNA were analyzed using a NanoDrop instrument (NanoDrop-1000, Wilmington, DE) and gel electrophoresis, respectively.
3.4. Designing of Primers
The sequences of ctxA, ToxR, and ompU genes of V. cholerae were obtained from Gene Bank, and their common areas were selected by Molecular Evolutionary Genetics Analysis 4 (MEGA4) software. To design the appropriate primers, allele ID and Gene Runner programs were applied. Finally, the specificity of each primer was investigated by performing BLAST in NCBI website. All the primers were constructed by Cinna Clon Co., Iran, as shown in Table 1.
Primers Sequences Designed for ctxA, toxR and ompU Genes; F and R Indicate the Forward and Reverse Primers, Respectively
| Target Gene | Primer Sequences 5΄ - 3΄ | Amplicon Size, bp | Source |
|---|---|---|---|
| Tox R-F | 5´- CCTTCGATCCCCTAAGCAATAC -3´ | 776 bp | original |
| Tox R-R | 5´- AGGGTTAGCAACGATGCGTAAG-3´ | ||
| Omp U-F | 5´-CGGTTGTCTAGGCTGTTGTTAG -3´ | 318 bp | original |
| Omp U -R | 5´- GACTCTGATTGCTCTTGCTGTAT-3´ | ||
| Ctx A-F | 5´- CAGAATGAGTACTTTGACCGAGG-3´ | 419 bp | original |
| Ctx A-R | 5´- GCCAATCCATAACCATCTGC -3´ |
3.5. Uniplex and Multiplex PCR Protocols
Three separate PCR reactions were accomplished for each designed primer in a final volume of 50 μL. The content of each reaction mixture included 5 μL of 10 X PCR buffer, 0.25 mM of each dNTP, 10 pM of each desired reverse and forward primer, 2 mM of MgCl2, 1 U of super Taq polymerase (Cinna Clon Co., Iran), and 50 ng of DNA template. The reaction mixture utilized for multiplex PCR was similar to the uniplex assay. The only discrepancy between them was the spontaneous addition of all three primers in the multiplex mixture.
PCR was performed in an Eppondorf thermal cycler with an initial denaturation step of 5 minutes at 94°C; 35 cycles of 1 minute at 94°C, 1 minute at 60°C, and 1 minute at 72°C, as well as a final extension step of 5 minutes at 72°C. Finally, the presence of amplified products was assessed using 1% agarose gel electrophoresis, stained by 0.5 ng/mL ethidium bromide and visualized under UV illumination. For further corroboration, relative bands from three different PCRs were extracted and purified from agarose gel (Cinagen, Iran) using the DNA extraction kit (Ferrmentas, Germany) and sequenced (Macrogen Research, Seoul, Korea). The abovementioned sequenced genes had local similarity with the V. cholerae ATCC 62013 standard strain, which was identified by BLAST; their homology was 99% as determined by MEGA4 software.
3.6. of Specificity and Sensitivity Evaluation of the Designed Primers
To analyze the specificity of the designed primers, 50 ng of the genomic DNA purified from standard species of V. cholerae ATCC 62013, S. typhi ATCC 14028, S. dysenteriae ATCC 13313 , and Escherichia coli O157:H7 ATCC 43894 were used for amplification by uniplex and multiplex tests. In addition, the sensitivity of the PCR tests was measured by preparing 10-fold serial dilutions of V. cholerae genome from 10 pg to 100 ng. PCR was performed for each concentration according to the protocol described in the previous section.
4. Results
4.1. Sample Collection
In the period of performing this study, from the total of stool samples collected, 72 V. cholerae isolates were differentiated by standard bacteriological tests described before. All of them were El Tor O1 strains.
4.2. Uniplex and Multiplex PCR Reactions
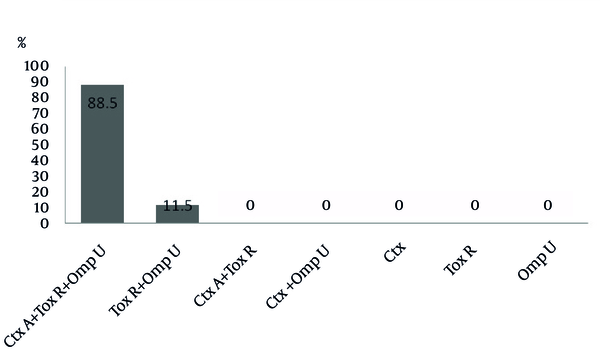
The presence of ctxA, toxR and ompU genes was assessed using specific primers for each gene, separately. The sizes of ctxA, toxR and ompU gene products were 419 base pair (bp), 776 bp, and 318 bp, respectively (Figure 1). All V. cholerae isolates separated from clinical samples possessed toxR and ompU genes. In addition, the presence of ctxA gene was detected in 61 (85%) of the isolates. Genotype frequencies of the isolates are illustrated in Figure 2.
Gel Electrophoresis of the PCR Products From ompU (318 bp), ctxA (419 bp), and ToxR (776 bp) Genes
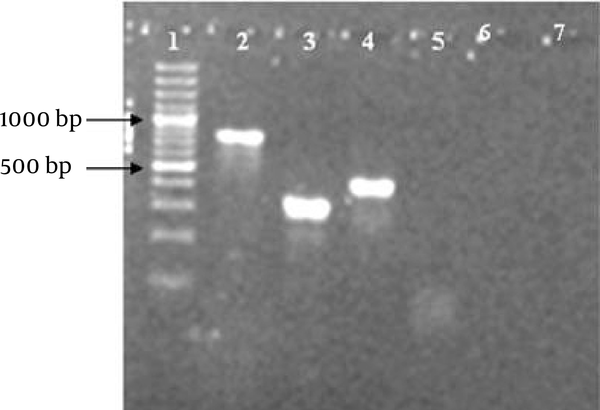
The Frequency of Different Genotypes Obtained in This Study

Results from the multiplex PCR were similar to uniplex PCR without any excessive products (as shown in Figure 3).
Gel Electrophoresis of ompU (318 bp), ctxA (419 bp), and ToxR (776 bp) Products Based on the Multiplex PCR Method
3.3. Specificity and Sensitivity Analyses
To verify the specificity of the designed primers for V. cholerae , purified genomic DNAs from S. typhi , S. dysenteriae and E. coli O157:H7 were tested. No product was seen (Figure 1). This result indicates the high specificity of constructed primers only for V. cholerae . Since V. cholerae is able to cause infection at the low count of 108 bacteria, the power of this test was analyzed in the range of 10 pg to 100 ng. Minimum detectable concentrations for ctxA, toxR and ompU genes in the applied PCR method were 10 pg, 10 pg and 50 pg, respectively.
5. Discussion
Diarrhea due to V. cholerae is an endemic disease in many developing countries. In the recent years, several outbreaks have been reported from Iran particularly within 2004 - 2009 (19). Despite the extensive attempts of health care communities and the related organizations to control and confine cholera, its incidence remains high (14). One of the most critical routes for preventing the outbreak of V. cholerae is the early stages diagnosis of the infection. Numerous microbiological tests including bacteria culture, serotyping, and biochemical methods have been developed; but most of them are time-consuming (20). Moreover, inaccurate results and lack of sensitivity in detection of low-dose infectious agents are disadvantages and limitations of the current methods.
Today, techniques based on nucleic acids such as PCR can be considered as rapid and reliable tests. In the recent years, uniplex PCR method has been developed to assess the toxigenicity potency of V. cholerae (5, 21), but its inability to simultaneously determine both toxigenic and regulatory factors has limited its usage. Multiplex PCR can provide a useful assay to eliminate this problem. In the study designed by Nandi and his colleagues, the presence of ompW and toxR genes was examined in 254 V. cholerae strains belonging to serogroups O1, O139, and non-O1/non-O139 by multiplex PCR. ompW and toxR genes were found to be positive in 223 and 229 isolates, respectively. In that test, identification of ctx gene as a potent factor for the V. cholerae toxigenicity was not included (22).
Panickar used DNA microarrays and multiplex PCR to detect pathogenic V. cholerae. In that study, 10 genes were selected to screen the different species belonging to the Vibrio family. Specific primers and probes against rrh and viuB were designed for V. vulnificus. tlh, tdh, trh, and ORF8 genes were employed for V. parahaemolyticus, beside that, for detection of V. cholerae, ompU, toxR, and tcpI genes were selected. Furthermore, El Tor and classical biotypes were screened by hlyA gene detection (23). In agreement with the present study, the specificity of their test was 100%. However, we embedded ctxA gene instead of tcpI, because tcpI gene cannot differentiate between toxigenic and non-toxigenic V. cholerae O1 and non-O139. Hoshino et al. developed a multiplex PCR based on the detection of ctxA and rfb genes.
That assay was capable of detecting toxigenic Vibrio O1 and O139 in stool samples with 100% sensitivity and 95% specificity (24). In another study, Mehrabadi and his co-worker designed a new multiplex PCR method based on three targets, ctxA, tcpA and ompW genes, for detecting toxigenic V. Cholerea. They reported that this developed method is able to detect 8.5 - 85 pg of genomic DNA and 10 - 100 colony-forming units of V. Cholerea. In addition, the specificity of this technique is 100% (11). In our previous study, we developed a multiplex PCR to detect hlyA, ctxB and tcpI as virulence and regulatory genes. The prevalence of hlyA, ctxB and tcpI genes in clinical samples was 94.7%, 90.8% and 92.1%, respectively (25).
In the present investigation, three different genes were selected based on their capability of detecting the pathogenicity and the toxigenicity at the same time. The existence of toxR gene is significant as a regulating agent for expression and function of several other genes, which are on turn responsible for the pathogenecity of V. cholerae (9, 26, 27). Furthermore, toxR gene does not horizontally get transferred from pathogenic strains to non-pathogenic ones (28); hence, it is specific to vibrionacea.
V. cholerae can adhere to the intestinal epithelial cells and be colonized in the presence of ompU gene. This event leads to the mucosal permeability intensification by disturbing the tight junctions (7). All of our isolates possessed both genes. However, the other Gram-negative bacteria used as negative controls cannot be detected by this assay. Our approach indicates that the constructed primers for ompU and toxR genes are capable of identifying pathogenic strains with specificity. Lack of ctxA gene was demonstrated in 11 of the isolates. This finding indicates that existence of the cholera toxin is not necessary for diarrhea and other different mechanisms can be involved in its occurrence. Significantly, our assay can detect V. cholerae at picogram levels. Despite several advantages of multiplex PCR which were described above, the complex formation and competition between primers are important disadvantages (17).
we conclude that, in addition to advantages such as simplicity, ease of use, speed and accuracy, the multiplex assay we designed can be utilized for simultaneous identification of pathogenic and toxigenic V. cholerae strains with high specificity and sensitivity; hence, it can be applied as an alternative to current bacteriological tests.
Acknowledgements
References
-
1.
World health organization. Cholera, 2009. Wkly Epidemiol Rec. 2010;2010(85):293-308.
-
2.
Provenzano D, Klose KE. Altered expression of the ToxR-regulated porins OmpU and OmpT diminishes Vibrio cholerae bile resistance, virulence factor expression, and intestinal colonization. Proc Natl Acad Sci U S A. 2000;97(18):10220-4. [PubMed ID: 10944196]. https://doi.org/10.1073/pnas.170219997.
-
3.
Rahmani F, Fooladi AA, Marashi SM, Nourani MR. Drug resistance in Vibrio cholerae strains isolated from clinical specimens. Acta Microbiol Immunol Hung. 2012;59(1):77-84. [PubMed ID: 22510289]. https://doi.org/10.1556/AMicr.59.2012.1.8.
-
4.
Davis BM, Waldor MK. Filamentous phages linked to virulence of Vibrio cholerae. Curr Opin Microbiol. 2003;6(1):35-42. [PubMed ID: 12615217].
-
5.
Marashi SM, Bakhshi B, Fooladi AA, Tavakoli A, Sharifnia A, Pourshafie MR. Quantitative expression of cholera toxin mRNA in Vibrio cholerae isolates with different CTX cassette arrangements. J Med Microbiol. 2012;61(Pt 8):1071-3. [PubMed ID: 22516130]. https://doi.org/10.1099/jmm.0.038752-0.
-
6.
Faruque SM, Albert MJ, Mekalanos JJ. Epidemiology, genetics, and ecology of toxigenic Vibrio cholerae. Microbiol Mol Biol Rev. 1998;62(4):1301-14. [PubMed ID: 9841673].
-
7.
Baudry B, Fasano A, Ketley J, Kaper JB. Cloning of a gene (zot) encoding a new toxin produced by Vibrio cholerae. Infect Immun. 1992;60(2):428-34. [PubMed ID: 1730472].
-
8.
Peterson KM, Mekalanos JJ. Characterization of the Vibrio cholerae ToxR regulon: identification of novel genes involved in intestinal colonization. Infect Immun. 1988;56(11):2822-9. [PubMed ID: 2902009].
-
9.
Matson JS, Withey JH, DiRita VJ. Regulatory networks controlling Vibrio cholerae virulence gene expression. Infect Immun. 2007;75(12):5542-9. [PubMed ID: 17875629]. https://doi.org/10.1128/IAI.01094-07.
-
10.
Tarr CL, Patel JS, Puhr ND, Sowers EG, Bopp CA, Strockbine NA. Identification of Vibrio isolates by a multiplex PCR assay and rpoB sequence determination. J Clin Microbiol. 2007;45(1):134-40. [PubMed ID: 17093013]. https://doi.org/10.1128/JCM.01544-06.
-
11.
Mehrabadi JF, Morsali P, Nejad HR, Imani Fooladi AA. Detection of toxigenic Vibrio cholerae with new multiplex PCR. J Infect Public Health. 2012;5(3):263-7. [PubMed ID: 22632601]. https://doi.org/10.1016/j.jiph.2012.02.004.
-
12.
Fukushima H, Tsunomori Y, Seki R. Duplex real-time SYBR green PCR assays for detection of 17 species of food- or waterborne pathogens in stools. J Clin Microbiol. 2003;41(11):5134-46. [PubMed ID: 14605150].
-
13.
Yamazaki W, Seto K, Taguchi M, Ishibashi M, Inoue K. Sensitive and rapid detection of cholera toxin-producing Vibrio cholerae using a loop-mediated isothermal amplification. BMC Microbiol. 2008;8:94. [PubMed ID: 18547441]. https://doi.org/10.1186/1471-2180-8-94.
-
14.
Mousavi SL, Nazarian S, Amani J, Rahgerdi AK. Rapid screening of toxigenic vibrio cholerae O1 strains from south Iran by PCR-ELISA. Iran Biomed J. 2008;12(1):15-21. [PubMed ID: 18392091].
-
15.
Huang J, Zhu Y, Wen H, Zhang J, Huang S, Niu J, et al. Quadruplex real-time PCR assay for detection and identification of Vibrio cholerae O1 and O139 strains and determination of their toxigenic potential. Appl Environ Microbiol. 2009;75(22):6981-5. [PubMed ID: 19767462]. https://doi.org/10.1128/AEM.00517-09.
-
16.
Chua AL, Elina HT, Lim BH, Yean CY, Ravichandran M, Lalitha P. Development of a dry reagent-based triplex PCR for the detection of toxigenic and non-toxigenic Vibrio cholerae. J Med Microbiol. 2011;60(Pt 4):481-5. [PubMed ID: 21183596]. https://doi.org/10.1099/jmm.0.027433-0.
-
17.
Pimenta FP, Hirata R, Jr, Rosa AC, Milagres LG, Mattos-Guaraldi AL. A multiplex PCR assay for simultaneous detection of Corynebacterium diphtheriae and differentiation between non-toxigenic and toxigenic isolates. J Med Microbiol. 2008;57(Pt 11):1438-9. [PubMed ID: 18927428]. https://doi.org/10.1099/jmm.0.2008/000414-0.
-
18.
Allen SD, Janda WM, Koneman EW, Schreckenberger PC. Overview - koneman's color atlas and textbook of diagnostic microbiology. 6 ed. Philadelphia, PA: Lippincott Williams & Wilkins; 2005.
-
19.
Marashi SM, Nasr Esfahani B, Tavakoli A, Moghim S, Pourshafie MR, Salehi M. Simultaneous Detection of Integrase and Antibiotic Resistance Genes within SXT Constin in Vibrio cholerae O1 El Tor Strains Isolated from Iran Using Multiplex-PCR. Iran J Basic Med Sci. 2012;15(3):885-9. [PubMed ID: 23493053].
-
20.
Singh DV, Isac SR, Colwell RR. Development of a hexaplex PCR assay for rapid detection of virulence and regulatory genes in Vibrio cholerae and Vibrio mimicus. J Clin Microbiol. 2002;40(11):4321-4. [PubMed ID: 12409420].
-
21.
Ramamurthy T, Pal A, Bag PK, Bhattacharya SK, Nair GB, Kurozano H, et al. Detection of cholera toxin gene in stool specimens by polymerase chain reaction: comparison with bead enzyme-linked immunosorbent assay and culture method for laboratory diagnosis of cholera. J Clin Microbiol. 1993;31(11):3068-70. [PubMed ID: 8263204].
-
22.
Nandi B, Nandy RK, Mukhopadhyay S, Nair GB, Shimada T, Ghose AC. Rapid method for species-specific identification of Vibrio cholerae using primers targeted to the gene of outer membrane protein OmpW. J Clin Microbiol. 2000;38(11):4145-51. [PubMed ID: 11060082].
-
23.
Panicker G, Call DR, Krug MJ, Bej AK. Detection of pathogenic Vibrio spp. in shellfish by using multiplex PCR and DNA microarrays. Appl Environ Microbiol. 2004;70(12):7436-44. [PubMed ID: 15574946]. https://doi.org/10.1128/AEM.70.12.7436-7444.2004.
-
24.
Hoshino K, Yamasaki S, Mukhopadhyay AK, Chakraborty S, Basu A, Bhattacharya SK, et al. Development and evaluation of a multiplex PCR assay for rapid detection of toxigenic Vibrio cholerae O1 and O139. FEMS Immunol Med Microbiol. 1998;20(3):201-7. [PubMed ID: 9566491].
-
25.
Imani Fooladi A, Iman Islamieh D, Hosseini Doust R, Karami A, Marashi S. Design of a multiplex pcr method for detection of toxigenicpathogenicin vibrio cholerae. Asian Pac J Trop Med. 2012;6(2):412-20.
-
26.
Bina J, Zhu J, Dziejman M, Faruque S, Calderwood S, Mekalanos J. ToxR regulon of Vibrio cholerae and its expression in vibrios shed by cholera patients. Proc Natl Acad Sci U S A. 2003;100(5):2801-6. [PubMed ID: 12601157]. https://doi.org/10.1073/pnas.2628026100.
-
27.
Stonehouse E, Kovacikova G, Taylor RK, Skorupski K. Integration host factor positively regulates virulence gene expression in Vibrio cholerae. J Bacteriol. 2008;190(13):4736-48. [PubMed ID: 18456804]. https://doi.org/10.1128/JB.00089-08.
-
28.
Covacci A, Falkow S, Berg DE, Rappuoli R. Did the inheritance of a pathogenicity island modify the virulence of Helicobacter pylori? Trends Microbiol. 1997;5(5):205-8. [PubMed ID: 9160510]. https://doi.org/10.1016/S0966-842X(97)01035-4.