Abstract
Background:
Klebseilla pneumoniae causes urinary tract infections, nosocomial pneumonia and intra-abdominal infections. Capsular antigens are considered to be the ultimate virulence determinants. Among 77 capsular serotypes of K. pneumoniae, serotypes K1 and K2 are the most virulent ones in humans.Objectives:
We designed a PCR method for detection of capsular serotypes K1 and K2 of K. pneumoniae using genes cps cluster wzc and orf10 which are required for biosynthesis of capsular polysaccharides in K1 and K2 types, respectively.Materials and Methods:
We collected 89 K. pneumoniae clinical isolates from the patients of Labafinejad Hospital located in Tehran. Clinical isolates were mostly chosen from urine. We used PCR technique to detect isolates possessing K1 and K2 capsular polysaccharides based on the wzc and orf10 genes which are required for biosynthesis of capsular polysaccharide in K1 and K2 types, respectively. The results of PCR were compared with those obtained by capsular serotyping using Quellung test.Results:
Of 89 isolates of K. pneumoniae tested by both techniques, 10 (11.2%) belonged to K1 and 13 (14.6%) belonged to K2 serotypes, respectively.Conclusions:
We found that these serotypes are probably important in clinical specimens. PCR was a simple and inexpensive tool for identification of K. pneumoniae clinical isolates of K1 and K2 geno-serotypes.Keywords
1. Background
Klebsiella pneumoniae subspecies pneumoniae is a common Gram-negative pathogen causing community and nosocomial infections, including pneumonia, urinary tract, septicaemia and wound infections (1, 2). Nosocomial infections developed by diverse K. pneumoniae strains may be considered as opportunistic, rather than true pathogens, since they mostly affect debilitated patients (3, 4). In addition, outbreak of multidrug resistant of Klebsiella spp. especially extended-spectrum beta lactamase (ESBL) has led the treatment to limited options in recent years (1). Although nosocomial infections of K. pneumoniae have occurred worldwide, some manifestations of community-acquired infection (namely, liver abscess and community-acquired pneumonia, meningitis and endophthalmitis) have been geographically restricted and occurred only in Taiwan and South Africa. There are two explanations for this difference in clinical manifestations which include host (such as alcoholism, access healthcare, etc.) and organism factors (the factors related to the organism is capsular serotypes) (5).
Community-acquired K. pneumoniae strains that have been reported along with bacteraemia and associated with high mortality in Taiwan, Europe, North America and Japan, possess Serotypes K1 and K2 (2-6). Capsular serotypes K1 and K2 are considered as predominant virulent strains of K. pneumoniae (7). Several studies of bacterial pathogenesis have reported that serotype K1; magA is the possible virulence factor for K. pneumoniae liver abscess (8, 9). Thus, PCR analysis of magA is a rapid and accurate method to detect capsular K1 strains (2-10). Moreover, prevalence of K2 isolates suggests a need for rapid detection of K2 serotype in addition to K1 (11). Wzc Protein was analyzed to be participating in reversible phosphorylation of tyrosine proteins. Most of these proteins are also involved in production of exopolysaccharides. Because exopolysaccharides are important virulence factors, a possible relationship between tyrosine phosphorylation and bacterial pathogenicity has been proposed (12).
2. Objectives
In this study, we designed a PCR method for detection of capsular serotypes K1 and K2 of K. pneumoniae using highly specific cps genes cluster, wzc and orf10 which are required for biosynthesis of capsular polysaccharide.
3. Materials and Methods
3.1. Bacterial Strains
We examined 89 K. pneumoniae clinical isolates from Labafinejad hospital located in Tehran. Clinical isolates were mostly from urine, some from pus, bile, mucus, trachea and wound. Prior to molecular-serotyping (PCR), all clinical isolates were biochemically identified by conventional bacteriology tests as detailed previously (13). Reference strains of K. pneumoniae AY762939 and K. pneumoniae D21242 were used as positive controls for PCR reactions of K1 and K2 serotypes respectively.
3.2. PCR Assay
PCR was used for detection of capsular K1 and K2 serotypes of K. pneumoniae. The DNA template was extracted using phenol and chloroform method. Primer pairs were designed from the specific sequence for the open reading frame (ORF)-10 regions in the cps gene clusters of K2 strains and wzc gene of K1 strains. Moreover, they were analyzed for hair-pin loop and primer dimer formation using Gene Runner software (Hasting software, Hastings Inc.). The specificities of these primer sets was confirmed by BLAST search in GenBank. Primers are; wzc forward, 5’-AGATAGAGGTGTATTGTCGC -3’ and wzc reverse, 5’- GAGCTCTATATGTTGGATGC -3’ for K1 strains and (ORF)-10 forward, 5’- TCATACTTGACAGAGGGAGTAG 3’ and, (ORF)-10 reverse, 5’- ACGATCGTTACAGTGACAAG -3’ for K2 strains.
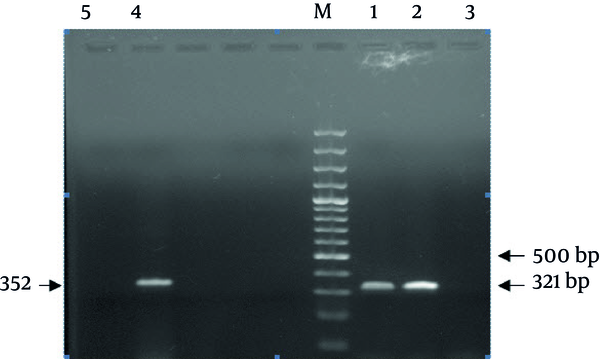
PCRs were carried out in 25 μL volumes containing 5 μL extracted DNA, with 1.5 U of Taq DNA polymerase (Fermentas Vilnius, Lithvania), 1X Mg-free PCR buffer, dNTP (Mix) at a concentration of 0.2 mM, 3.0 mM MgCl 2 , 0.8 μM of each Primer. Conditions were: 94°C for 5 minutes, followed by 35 cycles of 94°C for 45 seconds, 56°C for 45 seconds, 72°C for 45 seconds, and a final extension at 72°C for 5 minutes. The 2% agarose gel in TBE buffer was used for PCR products separation. Gels were run at a constant voltage of 100 V for 1 hour, stained in 2 μg/mL ethidium bromide for 10 minutes and photographed under UV by Gel-Document. The expected PCR products of (ORF)-10 and wzc were 321 and 352 base pair (bp) in length, respectively (Figure 1).
Result of the PCR Assay for Identification of K. pneumoniae Capsular K1 and K2
3.3. Quellung Test
The antisera against capsular antigens K1 and K2 of K. pneumoniae were purchased from Statens Serum Institu (Statens seum institute, Copenhagen, Denmark). The Quelling test was performed according to the instruction given by Edmandson and Cook (14).
4. Results
A total of 89 clinical isolates of K. pneumoniae [urine (n = 70), blood (n = 3), ulcer (n = 5), trachea (n = 6), sputum (n = 2) and Stool (n = 3)] were included in the study which had been obtained from Labafinejad hospital patients. Among 10 positive K1 serotype isolates, nine were urine samples and one was trachea. Also among 13 K2 serotypes isolates, 11 were urine and two were trachea samples. We developed a PCR technique to detect isolates possessing K1 and K2 capsular polysaccharides using wzc and orf10 specific genes which are required for biosynthesis of capsular polysaccharide types K1 and K2. Two 352 bp and 321 bp bands were observed for K1 and K2 reference strains, respectively. No band was observed in DNA-free negative control. Moreover, the developed assay correctly identified 10 isolates (11.2%) as K1 and 13 isolates (14.6%) as K2 serotypes among 89 isolates of K. pneumoniae serotypes. The results of PCR were confirmed by Quellung test.
5. Discussion
K. pneumoniae is an enterobacterial and opportunistic pathogen that causes urinary tract infection (UTI), pneumonia and wound infections. In this study, patients were mostly infected by urinary K1 and K2 serotypes, but in other studies patients were more likely to have liver abscesses and less likely to have UTIs or biliary tract infections (15). For the PCR assay, we selected wzc gene, encoding tyrosine-protein kinase and open reading frame 10 (orf-10) encoding putative inner membrane proteins for K1 and K2 serotypes, respectively. There have been many reports linking magA, rmpA and wcaG with virulence, also wzc and orf-10 have been studied (16).
Wzc protein was analyzed to evaluate K1 and K2 serotypes capacity to participate in the reversible phosphorylation of proteins on tyrosine. Wzc was found in other bacterial species which are all involved in the synthesis or export of exopolysaccharides. Since these are considered as important virulence factors, it could be suggested that reversible protein phosphorylation on tyrosine may be part of the cascade of reactions that determine the pathogenicity of bacteria (17). The open reading frames (ORFs) of the complete cps regions suggested that this region was responsible for capsular polysaccharide synthesis (16).
Our results suggest that PCR analysis is a rapid and reliable method for identification of both capsular K1 and K2 serotypes of K. pneumoniae. However, the techniques most commonly used for identification of K. pneumoniae serotypes K1 and K2 (Quellung and counter-current immunoelectrophoresis) are limited, because of costs of various antisera preparation. Thus, PCR assay may help to operate K. pneumoniae capsular K1 and K2 type identification in routine diagnoses. Great advantage of molecular serotyping is that it does not undergo cross-reactions which sometimes render highly ambiguous results for conventional serotyping (5). Therefore, PCR genotyping seems to be a more sensitive and specific way for detecting these serotypes (16). Furthermore, molecular serotyping is capable to determine a potential serotype of capsule-deficient isolates (7, 18, 19). We recommend that PCR analysis could be helpful in seroepidemiologic studies and early-tracing of contingent metastatic infections in patients with liver abscess (7, 10, 11, 15-20). This is the first report on the occurrence of serotypes K1 and K2 in the clinical specimens and shows the pathogenic potential of these isolates in different organs. Molecular-serotyping with PCR for serotypes K1 and K2 had not been previously studied in Iran. This may provide a new way to “genotype” an unknown capsular type strain but will require validation with several strains of each serotype to determine specificity and sensitivity.
Acknowledgements
References
-
1.
Podschun R, Ullmann U. Klebsiella spp. as nosocomial pathogens: epidemiology, taxonomy, typing methods, and pathogenicity factors. Clin Microbiol Rev. 1998;11(4):589-603. [PubMed ID: 9767057].
-
2.
Turton JF, Englender H, Gabriel SN, Turton SE, Kaufmann ME, Pitt TL. Genetically similar isolates of Klebsiella pneumoniae serotype K1 causing liver abscesses in three continents. J Med Microbiol. 2007;56(Pt 5):593-7. [PubMed ID: 17446279]. https://doi.org/10.1099/jmm.0.46964-0.
-
3.
Brisse S, Fevre C, Passet V, Issenhuth-Jeanjean S, Tournebize R, Diancourt L, et al. Virulent clones of Klebsiella pneumoniae: identification and evolutionary scenario based on genomic and phenotypic characterization. PLoS One. 2009;4(3). [PubMed ID: 19319196]. https://doi.org/10.1371/journal.pone.0004982.
-
4.
Lau HY, Clegg S, Moore TA. Identification of Klebsiella pneumoniae genes uniquely expressed in a strain virulent using a murine model of bacterial pneumonia. Microb Pathog. 2007;42(4):148-55. [PubMed ID: 17369011]. https://doi.org/10.1016/j.micpath.2007.01.001.
-
5.
Yu VL, Hansen DS, Ko WC, Sagnimeni A, Klugman KP, von Gottberg A, et al. Virulence characteristics of Klebsiella and clinical manifestations of K. pneumoniae bloodstream infections. Emerg Infect Dis. 2007;13(7):986-93. [PubMed ID: 18214169]. https://doi.org/10.3201/eid1307.070187.
-
6.
Diancourt L, Passet V, Verhoef J, Grimont PA, Brisse S. Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. J Clin Microbiol. 2005;43(8):4178-82. [PubMed ID: 16081970]. https://doi.org/10.1128/JCM.43.8.4178-4182.2005.
-
7.
Turton JF, Baklan H, Siu LK, Kaufmann ME, Pitt TL. Evaluation of a multiplex PCR for detection of serotypes K1, K2 and K5 in Klebsiella sp. and comparison of isolates within these serotypes. FEMS Microbiol Lett. 2008;284(2):247-52. [PubMed ID: 18507682]. https://doi.org/10.1111/j.1574-6968.2008.01208.x.
-
8.
Fang CT, Lai SY, Yi WC, Hsueh PR, Liu KL, Chang SC. Klebsiella pneumoniae genotype K1: an emerging pathogen that causes septic ocular or central nervous system complications from pyogenic liver abscess. Clin Infect Dis. 2007;45(3):284-93. [PubMed ID: 17599305]. https://doi.org/10.1086/519262.
-
9.
Yu WL, Fung CP, Ko WC, Cheng KC, Lee CC, Chuang YC. Polymerase chain reaction analysis for detecting capsule serotypes K1 and K2 of Klebsiella pneumoniae causing abscesses of the liver and other sites. J Infect Dis. 2007;195(8):1235-6. author reply 1236. [PubMed ID: 17357063]. https://doi.org/10.1086/512686.
-
10.
Chuang YP, Fang CT, Lai SY, Chang SC, Wang JT. Genetic determinants of capsular serotype K1 of Klebsiella pneumoniae causing primary pyogenic liver abscess. J Infect Dis. 2006;193(5):645-54. [PubMed ID: 16453259]. https://doi.org/10.1086/499968.
-
11.
Cryz SJ, Jr, Mortimer PM, Mansfield V, Germanier R. Seroepidemiology of Klebsiella bacteremic isolates and implications for vaccine development. J Clin Microbiol. 1986;23(4):687-90. [PubMed ID: 3517058].
-
12.
Grangeasse C, Doublet P, Cozzone AJ. Tyrosine phosphorylation of protein kinase Wzc from Escherichia coli K12 occurs through a two-step process. J Biol Chem. 2002;277(9):7127-35. [PubMed ID: 11751920]. https://doi.org/10.1074/jbc.M110880200.
-
13.
Feizabadi MM, Delfani S, Raji N, Majnooni A, Aligholi M, Shahcheraghi F, et al. Distribution of bla(TEM), bla(SHV), bla(CTX-M) genes among clinical isolates of Klebsiella pneumoniae at Labbafinejad Hospital, Tehran, Iran. Microb Drug Resist. 2010;16(1):49-53. [PubMed ID: 19961397]. https://doi.org/10.1089/mdr.2009.0096.
-
14.
Edmondson AS, Cooke EM. The production of antisera to the Klebsiella capsular antigens. J Appl Bacteriol. 1979;46(3):579-84. [PubMed ID: 479055].
-
15.
Liao CH, Huang YT, Lai CC, Chang CY, Chu FY, Hsu MS, et al. Klebsiella pneumoniae bacteremia and capsular serotypes, Taiwan. Emerg Infect Dis. 2011;17(6):1113-5. [PubMed ID: 21749784]. https://doi.org/10.3201/eid/1706.100811.
-
16.
Pan YJ, Fang HC, Yang HC, Lin TL, Hsieh PF, Tsai FC, et al. Capsular polysaccharide synthesis regions in Klebsiella pneumoniae serotype K57 and a new capsular serotype. J Clin Microbiol. 2008;46(7):2231-40. [PubMed ID: 18508935]. https://doi.org/10.1128/JCM.01716-07.
-
17.
Vincent C, Doublet P, Grangeasse C, Vaganay E, Cozzone AJ, Duclos B. Cells of Escherichia coli contain a protein-tyrosine kinase, Wzc, and a phosphotyrosine-protein phosphatase, Wzb. J Bacteriol. 1999;181(11):3472-7. [PubMed ID: 10348860].
-
18.
Gierczynski R, Jagielski M, Rastawicki W, Kaluzewski S. Multiplex-PCR assay for identification of Klebsiella pneumoniae isolates carrying the cps loci for K1 and K2 capsule biosynthesis. Pol J Microbiol. 2007;56(3):153-6. [PubMed ID: 18062647].
-
19.
Brisse S, Issenhuth-Jeanjean S, Grimont PA. Molecular serotyping of Klebsiella species isolates by restriction of the amplified capsular antigen gene cluster. J Clin Microbiol. 2004;42(8):3388-98. [PubMed ID: 15297473]. https://doi.org/10.1128/JCM.42.8.3388-3398.2004.
-
20.
Struve C, Bojer M, Nielsen EM, Hansen DS, Krogfelt KA. Investigation of the putative virulence gene magA in a worldwide collection of 495 Klebsiella isolates: magA is restricted to the gene cluster of Klebsiella pneumoniae capsule serotype K1. J Med Microbiol. 2005;54(Pt 11):1111-3. [PubMed ID: 16192445]. https://doi.org/10.1099/jmm.0.46165-0.