Abstract
Background:
The number of invasive candidiasis cases has risen especially with an increase in the number of immunosuppressed and immunocom promised patients. The early detection of Candida species which is specific and sensitive is important in determining the correct administration of antifungal drugs to patients.Objectives:
This study aims to develop a method for the detection, identification and quantitation of medically important Candida species through quantitative polymerase chain reaction (qPCR).Materials and Methods:
The isocitrate lyase (ICL) gene which is not found in mammals was chosen as the target gene of real-time PCR. Absolute quantitation of the gene copy number was achieved by constructing the plasmid containing the ICL gene which is used to generate standard curve. Twenty fungal species, two bacterial species and human DNA were tested to check the specificity of the detection method.Results:
All eight Candida species were successfully detected, identified and quantitated based on the ICL gene. A seven-log range of the gene copy number and a minimum detection limit of 103 copies were achieved.Conclusions:
A one-tube absolute quantification real-time PCR that differentiates medically important Candida species via individual unique melting temperature was achieved. Analytical sensitivity and specificity were not compromised.Keywords
1. Background
There has been an increase of invasive candidiasis cases caused by non-albicans Candida species compared to Candida albicans over the last three decades (1). These species presented different drug susceptibility patterns towards current and established antifungals (2). Therefore, there is an urgent need to accurately identify the species for a proper management of patients.
2. Objectives
This study aims to develop a specific and sensitive absolute quantitation real-time PCR (qPCR) method medically targeting the important Candida species. We detected common homologous regions in the isocitrate lyase gene and also in Candida species and applied them as target for primers design.
3. Materials and Methods
Isocitrate lyase gene sequences in C. albicans (Accession No.: AF222905), C. tropicalis (Accession No.: D00703) and C. utilis (Accession No.: AJ404885) were aligned using the software CLUSTALW(is a free software) (3) and the forward primer, ICL1c 5’- GAC CAC AGA GAC CA TAC TT- 3’ and reverse primer, ICLr 5’- TGA AAG ATG GRG ACA AGT T- 3’ respectively were designed. All PCR primers were synthesized by the First BASE Laboratories Sdn. Bhd., Malaysia. For the standard curve employed in the absolute quantification, full length sequence of C. albicans isocitrate lyase gene was cloned by PCR and inserted into p-GEMT Easy Vector (Promega, USA). The full length of the isocitrate lyase gene of C. albicans (Accession No.: AF222905) was retrieved from the GenBank. PCR primers were designed; ICLflF 5’-ACC ATG CCT TAC ACT CCT ATT GAC A- 3’ and ICLflR 5’- TTA AGC CTT GGC TTT GGA TTC TTT G - 3’. PCR was performed at a 20-µL final volume containing 1X reaction buffer [(10 mM Tris-HCl (pH 8.8), 50 mM KCl, 0.08% Nonidet P40 (Fermentas Life Sciences, Canada)], 2.0 mM MgCl2, 0.2 mM dNTPs, 0.5 µM forward and reverse primers (ICLf1F and ICLf1R) and 1 U Taq DNA polymerase (Fermentas Life Sciences, Canada) to amplify the gene and template DNA was extracted from C. albicans ATCC 14053.
The PCR cycle was as follow: incubation at 94°C for two minutes, followed by 40 amplification cycles of 94°C for 30 s, 58.4°C for 30 s, 72°C for 1.5 minutes with final extension cycle at 72°C for 4 minutes. The amplified product (1653 bp) was confirmed through 1% agarose gel electrophoresis. The amplified products were purified using QIA quick Gel Extraction Kit (QIAGEN, Germany) through gel purification method. Cloning was performed in 10 µL of volume containing 1 X Rapid Ligation Buffer [(60 mM Tris-HCL) pH 7.8), 20 mM MgCl2, 20 mm DTT, 2 mM ATP, 10% polyethylene glycol)], 50 ng of pGEM®-T Easy vector (Please refer Supplementary materials for the vector map), 3 µL of purified PCR product (80 ng) and 3 Weiss unit of T4 DNA ligase and the mixture was incubated at 4°C overnight.
The overnight ligation mixture was mixed with TOP 10 E. coli competent cells and transformed and plated onto DifcoTM LB agar (Becton Dickinson, USA) plates containing 100 µg/mL ampicillin, 100 mM IPTG and 40 mg/mL X-gal, and incubated at 37°C overnight at. The positive clones were picked and subcultured in DifcoTM LB broth (Becton Dickinson, USA) and the vector containing the isocitrate lyase gene insert was isolated and confirmed through DNA sequencing. The recombinant plasmid vectors with isocitrate lyase gene insert were now ready to be used for the qPCR testing. Prior to that, the plasmid were linearised using the restriction enzyme PstI (Fermentas, Lithuania), purified and calculated based on the formula found in the Handbooks and Protocols, Qiagen to find the copy number of the plasmid. The qPCR amplification was performed in Rotor-Gene 3000 (Corbett Research, Australia) and all PCR reagents were supplied from Fermentas Life Sciences (Canada).
The PCR reaction was performed in a final volume of 20 µL. The PCR mixture contained 1 X PCR buffer (10 mM Tris-HCl, pH 8.8, 50 mM KCl, 0.08% Nonidet P40), 2.25 mM of MgCl2, 0.25 mM each deoxynucleoside triphosphates (dNTPs), 0.25 μM of each forward and reverse primers, 0.5 U of Taq DNA polymerase, 0.5 L of 10 X SYBR Green dye (Cambrex, USA) and DNA/plasmid template. The PCR cycle included: incubation at 95°C for 3 minutes, followed by 34 amplification cycles of 94°C for 20 s, 50°C for 20 s, 72°C for 20 s and 15 s at 80°C (for fluorescence acquiring). For melting curve analysis, the temperature was set to rise from 80°C to 99°C with a 45-second-hold on the first step and a 5-second-hold on every next step.
Amplicons were sequenced to confirm the DNA sequence of the expected species. In order to scrutinize the analytical specificity of the method, a list of ATCC (ATCC, USA) and clinical isolates strains of various fungal and bacterial species together with seven genomic DNA from an apparently healthy human volunteer were tested (Table 1). Prior to testing, species of the clinical fungal isolates were identified at the Microbiology Diagnostic Lab, Universiti Malaya Medical Center, Kuala Lumpur (4). Normal human volunteer blood was taken under the human ethics committee approval and all the participants singed an informed consent form. Fungal DNA was extracted using modified conventional phenol chloroform extraction method (5). Human and bacterial DNA were extracted using GENEALL DNA extraction kit (General Biosystem, Korea). For analytical sensitivity testing, we used a 10 X serially diluted recombinant plasmid containing the isocitrate lyase gene, which was earlier constructed.
The ATCC Reference Strains and Clinical Isolates Used in the Analytical Specificity Testinga
| Fungi Species | Total |
|---|---|
| Candida species | 30 |
| C. albicans | 5 |
| C. dubliniensis | 3 |
| C. guilliermondii | 1 |
| C. kefyr | 1 |
| C. krusei | 9 |
| C. lusitaniae | 1 |
| C. parapsilosis | 5 |
| C. tropicalis | 5 |
| Aspergillus and Cryptococcus species | 22 |
| A. flavus | 4 |
| A. fumigatus | 3 |
| A. lentulus | 1 |
| A. nidulans | 1 |
| A. niger | 3 |
| A. nomius | 2 |
| A. sojae | 1 |
| A. tamarii | 2 |
| Other bacteria | 4 |
| Escherichia coli | 2 |
| Staphyloccoccus aureus | 2 |
| Other organism | 7 |
| Homo sapiens | 7 |
4. Results
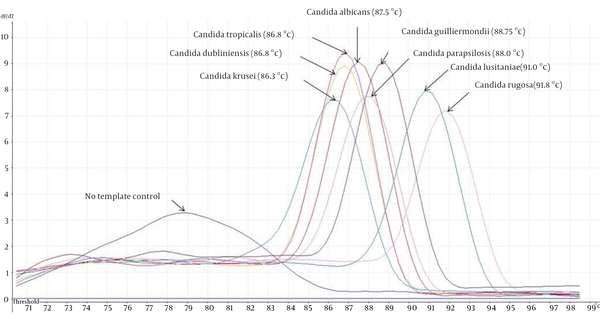
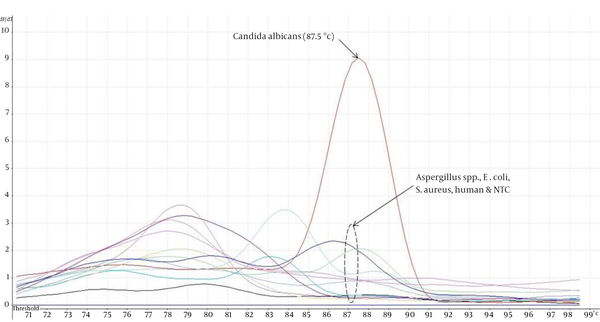
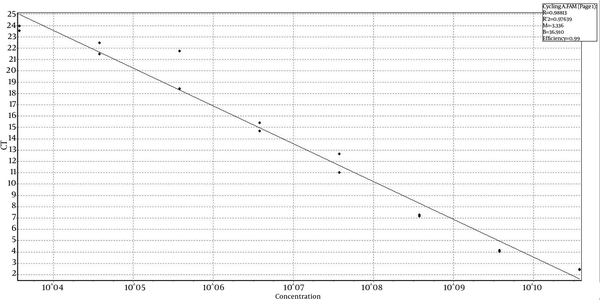
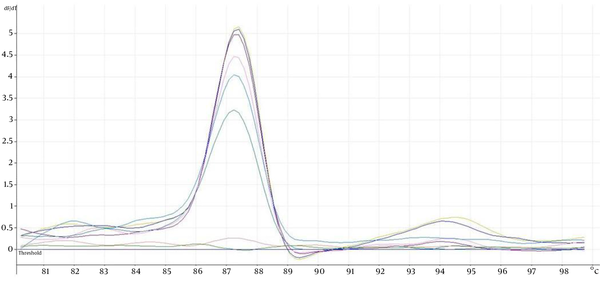
The eight ATCC Candida species showed specific unique individual melting temperature value respectively except for C. dubliniensis and C. tropicalis (Figure 1). DNA of the other medically important fungi and bacteria together with human genomic DNA showed no amplification (Supplementary materials). Twenty-two clinical Candida isolates of various species were further tested and all recorded the expected melting temperature values as shown by their respective ATCC species. The summary of the analytical specificity testing is found in Table 1. For the analytical sensitivity, a seven-log range of the copy number of isocitrate lyase gene and a minimum detection limit of 103 copies of isocitrate lyase gene were achieved (Supplementary materials).
Melting Curve Analysis of Different Candida ATCC With Their Respective Melting Temperature in Parenthesis
5. Discussion
In comparison to conventional PCR, the qPCR has added advantages over the former ones (6). Previous studies which utilized other PCR platforms such as semi nested PCR (4) and nested PCR (7) fall short in simplifying the detection process where it requires the use of multiple tubes. In this study, a single tube detection of eight medically important Candida species were successfully achieved through their unique product melting temperature as opposed to five species reported in the previous study (8). Exception was seen on two of the species namely C. dubliniensis and C. tropicalis that both shared similar profile (Figure 1). This scenario prompted us to sequence the PCR amplicons to rule out cross-contamination. Both nucleotide sequences were found to be unique themselves and the homology was 86% after BLAST and multiple sequence alignment analysis.
To differentiate both species in such situation, we propose a simple germ tube test in which the C. dubliniensis provides positive results (9) while the C. tropicalis does not. This study was able to determine the exact copy number of the gene present (fungal load) with a relatively wider dynamic range of seven-log level. This was an additional two-log range in comparison to the previously reported study on Candida detection (8). A good detection method is not just useful for detection per se but also for the quantitation with which the findings can be useful in knowing the dynamics of pathogen proliferation, monitoring of response to treatment and also helping to avoid false positive results when used constitutively in monitoring (10).
Isocitrate lyase is one of the genes that are involved in the glyoxylate cycle, of which it is not found in mammals (11). This renders a high possibility that this gene might be used as a potential target for Candida species detection without potential false positive issues related to cross-amplification of human DNA samples. In addition to the main aim of this study, the partial sequence of isocitrate lyase gene of other Candida species were successfully sequenced and consequently deposited in GenBank; Kluyveromyces marxianus (anamorph C. kefyr) Accession No.: GQ922892, Clavispora lusitaniae (anamorph C. lusitaniae) Accession No.: GQ922890.
Melting curve analysis of Candida albicans, six Aspergillus species ATCC strains, Escherichia coli, Staphylococcus aureus, human and no template control (NTC).
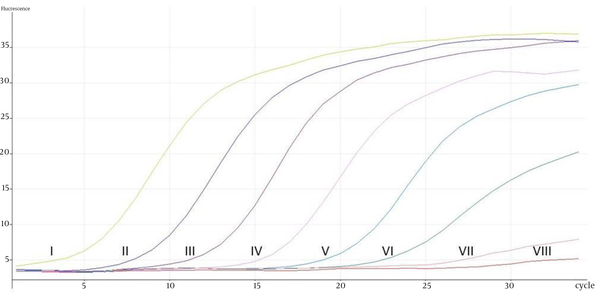
Representative Real-Time PCR Cycling Profiles of Isocitrate Lyase Gene From pGEM-T Isocitrate Lyase Gene Plasmid DNA With Seven-Log Range Concentration Ranging From 3.77 × 103 to 3.77 × 1010 Copies; I- 3.77 × 1010 copies,
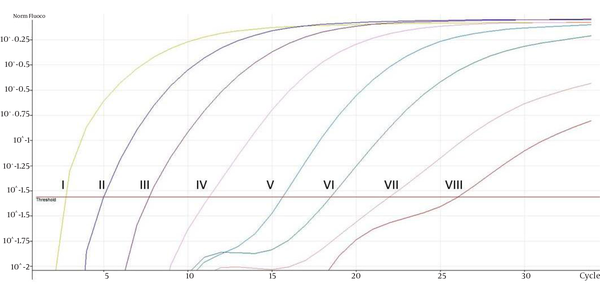
Representative Real-Time Quantification Curve for Isocitrate Lyase Gene With Seven-Log Range Concentrations Ranging From 3.77 × 103 to 3.77 × 1010 Copies; I- 3.77 × 1010 copies, II- 3.77 × 109 copies, III- 3.77
Representative Graph Showing Seven-Log Dynamic Range of 3.77 x 103 to 3.77 x 1010 Copies Through a Series of Ten Times Serial Dilution of pGEM-T Isocitrate Lyase Gene
Representative Melting Curves Analysis for Isocitrate Lyase Gene Amplification Products From pGEM-T Isocitrate Lyase Gene Plasmid DNA of Eight Concentrations, 3.77 × 103 to 3.77 × 1010 Copies With the Melting Temperature at 87.5°C
pGEM®-T Easy Vector Map and Sequence Reference Points (Adapted From the Promega Technical Manual of pGEM®-T and pGEM®-T Easy Vectors Systems)
Acknowledgements
References
-
1.
Diekema D, Arbefeville S, Boyken L, Kroeger J, Pfaller M. The changing epidemiology of healthcare-associated candidemia over three decades. Diagn Microbiol Infect Dis. 2012;73(1):45-8. [PubMed ID: 22578938]. https://doi.org/10.1016/j.diagmicrobio.2012.02.001.
-
2.
Pfaller MA, Diekema DJ, Gibbs DL, Newell VA, Ellis D, Tullio V, et al. Results from the ARTEMIS DISK Global Antifungal Surveillance Study, 1997 to 2007: a 10.5-year analysis of susceptibilities of Candida Species to fluconazole and voriconazole as determined by CLSI standardized disk diffusion. J Clin Microbiol. 2010;48(4):1366-77. [PubMed ID: 20164282]. https://doi.org/10.1128/JCM.02117-09.
-
3.
Subramaniam S. The Biology Workbench--a seamless database and analysis environment for the biologist. Proteins. 1998;32(1):1-2. [PubMed ID: 9672036].
-
4.
Than LT, Chong PP, Ng KP, Seow HF. Detection of 10 medically important Candida species by seminested polymerase chain reaction. Diagn Microbiol Infect Dis. 2012;72(2):196-8. [PubMed ID: 22154674]. https://doi.org/10.1016/j.diagmicrobio.2011.10.008.
-
5.
Buchman TG, Rossier M, Merz WG, Charache P. Detection of surgical pathogens by in vitro DNA amplification. Part I. Rapid identification of Candida albicans by in vitro amplification of a fungus-specific gene. Surgery. 1990;108(2):338-46. [PubMed ID: 2200156].
-
6.
Klein D. Quantification using real-time PCR technology: applications and limitations. Trends Mol Med. 2002;8(6):257-60. [PubMed ID: 12067606].
-
7.
Jaeger EE, Carroll NM, Choudhury S, Dunlop AA, Towler HM, Matheson MM, et al. Rapid detection and identification of Candida, Aspergillus, and Fusarium species in ocular samples using nested PCR. J Clin Microbiol. 2000;38(8):2902-8. [PubMed ID: 10921948].
-
8.
Dunyach C, Bertout S, Phelipeau C, Drakulovski P, Reynes J, Mallie M. Detection and identification of Candida spp. in human serum by LightCycler real-time polymerase chain reaction. Diagn Microbiol Infect Dis. 2008;60(3):263-71. [PubMed ID: 18036761]. https://doi.org/10.1016/j.diagmicrobio.2007.09.014.
-
9.
Sullivan DJ, Westerneng TJ, Haynes KA, Bennett DE, Coleman DC. Candida dubliniensis sp. nov.: phenotypic and molecular characterization of a novel species associated with oral candidosis in HIV-infected individuals. Microbiology. 1995;141 ( Pt 7):1507-21. [PubMed ID: 7551019].
-
10.
Watzinger F, Ebner K, Lion T. Detection and monitoring of virus infections by real-time PCR. Mol Aspects Med. 2006;27(2-3):254-98. [PubMed ID: 16481036]. https://doi.org/10.1016/j.mam.2005.12.001.
-
11.
Lorenz MC, Fink GR. The glyoxylate cycle is required for fungal virulence. Nature. 2001;412(6842):83-6. [PubMed ID: 11452311]. https://doi.org/10.1038/35083594.